View data
import numpy as np import pandas as pd import matplotlib.pyplot as plt
data=pd.read_csv("code/ex2-logistic regression/ex2data1.txt",names=['Exam 1', 'Exam 2', 'Admitted']) data.head()

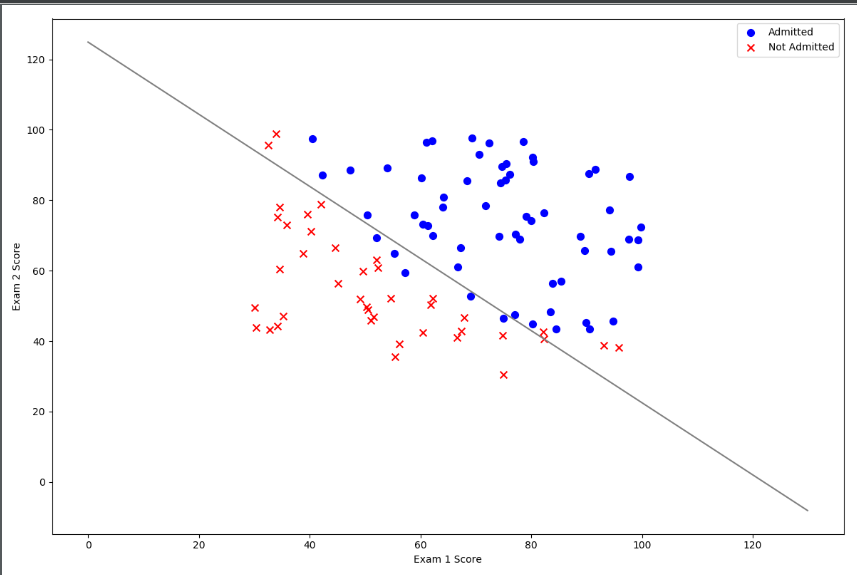
positive=data[data["Admitted"].isin(["1"])] negative=data[data["Admitted"].isin(["0"])] fig,ax=plt.subplots(figsize=[12,8]) ax.scatter(positive['Exam 1'], positive['Exam 2'], s=50, c='b', marker='o', label='positive') ax.scatter(negative['Exam 1'], negative['Exam 2'], s=50, c='r', marker='x', label='negative') ax.legend() ax.set_xlabel('Exam 1 Score') ax.set_ylabel('Exam 2 Score') plt.show()
About pandas.isin() A kind of :
isin() accepts a list and determines whether the elements in the column are in the list.
Give an example:
>>> df
A B C D E
0 -0.018330 2.093506 -0.086293 -2.150479 a
1 0.104931 -0.271810 -0.054599 0.361612 a
2 0.590216 0.218049 0.157213 0.643540 c
3 -0.254449 -0.593278 -0.150455 -0.244485 b
>>> df.E.isin(['a','c'])
0 True
1 True
2 True
3 False
Name: E, dtype: bool
--------
Copyright notice: This is the original article of CSDN blogger "lzw2016", following CC 4.0 BY-SA copyright agreement. Please attach the original source link and this notice for reprint.
Original link: https://blog.csdn.net/lzw2016/article/details/80472649
After data visualization:
sigmoid function
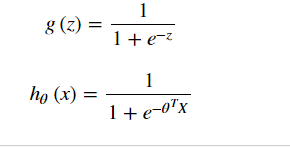
def sigmoid(z): return 1/(1+np.exp(-z))
Cost function
For linear regression models, the error we define is the sum of the squares of all model errors: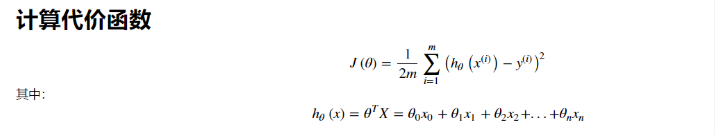
But for the logistic regression model, the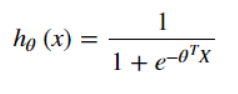 By substituting the above cost function, we will get a nonconvex function, which leads to many local minima in our cost function, which will affect the gradient descent algorithm to find the global minima.
By substituting the above cost function, we will get a nonconvex function, which leads to many local minima in our cost function, which will affect the gradient descent algorithm to find the global minima.
So we redefine the cost function: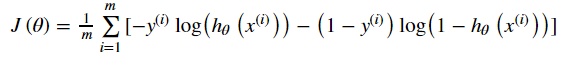
data.insert(0,"ones",1)#Add a column x0=1 so that the number of X is the same as theta cols=data.shape[1] X=data.iloc[:,0:cols-1] Y=data.iloc[:,cols-1:cols] X=np.array(X.values) Y=np.array(Y.values) theta=np.zeros(3)
def cost(theta,X,Y): theta=np.matrix(theta) z=np.dot(X,theta.T) m=len(X) cost=1/m*np.sum(np.multiply(-Y,np.log(sigmoid(z)))-np.multiply((1-Y),np.log(1-sigmoid(z)))) return cost
cost(theta,X,Y)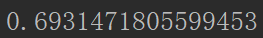
theta=np.matrix(theta) changes theta from (3,) to (3, 1)
The former is one-dimensional, the latter is two-dimensional.
For example:
The result of NP. Sum (two-dimensional matrix, axis=1) is one-dimensional
NP. Sum (two-dimensional matrix, axis=1,keepdims=True) results in two-dimensional
On the difference between Python list, Numpy array and matrix
gradient descent
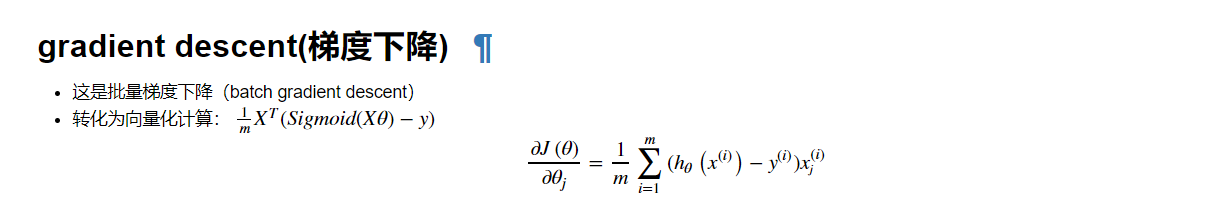
def gradient(theta,X,Y): theta = np.matrix(theta) X = np.matrix(X) Y = np.matrix(Y) parameters=int(theta.ravel().shape[1]) grads=np.zeros(parameters) z=np.dot(X,theta.T) error=sigmoid(z)-Y for i in range(parameters): term=np.multiply(error,X[:,i]) grads[i]=np.sum(term)/len(X) return grads gradient(theta,X,Y)

Note that we are not actually performing gradient descent in this function, we are only calculating a gradient step. In the exercise, an Octave function called "fminunc" is used to optimize the function to calculate the cost and gradient parameters. Because we use Python, we can do the same with SciPy's "optimize" namespace.
import scipy.optimize as opt res = opt.minimize(fun=cost, x0=theta, args=(X, Y), method='Newton-CG', jac=gradient) print(res)
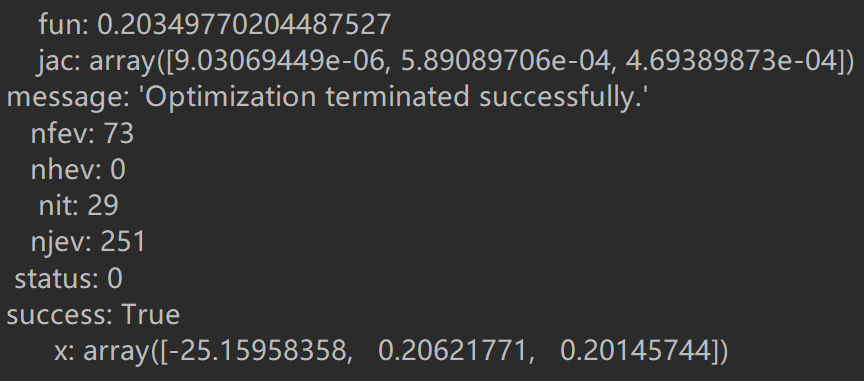
About scipy.optimize:
scipy.optimize.minimize(fun, x0, args=(), method=None, jac=None, hess=None, hessp=None, bounds=None, constraints=(), tol=None, callback=None, options=None)[source]¶
- fun: minimized objective function
- x0: initialized parameters
- args: parameters passed to the target function
- Optimization method:

- bounds: limits on variables for l-bfgs-b, TNC, slsqp and
trust-constr methods.) - Constraints: Constraints definition (only for COBYLA, SLSQP and
trust-constr)
......
scipy.optimize.minimize
Forecast

def predict(theta, X): z=np.dot(X,theta.T) probs = sigmoid(z) return [1 if x >= 0.5 else 0 for x in probs]
theta_min = np.matrix(res.x) predictions = predict(theta_min, X) correct = [1 if ((a == 1 and b == 1) or (a == 0 and b == 0)) else 0 for (a, b) in zip(predictions, Y)] accuracy = (sum(map(int, correct)) % len(correct)) print ('accuracy = {0}%'.format(accuracy))
accuracy = 89%
Drawing decision boundaries
coef = -(res.x / res.x[2]) # find the equation print(coef) x = np.arange(130, step=0.1) y = coef[0] + coef[1]*x data.describe() # find the range of x and y
coef[0] is intercept, about 125
fig, ax = plt.subplots(figsize=(12,8)) ax.scatter(positive['Exam 1'], positive['Exam 2'], s=50, c='b', marker='o', label='Admitted') ax.scatter(negative['Exam 1'], negative['Exam 2'], s=50, c='r', marker='x', label='Not Admitted') ax.plot(x,y,'grey') ax.legend() ax.set_xlabel('Exam 1 Score') ax.set_ylabel('Exam 2 Score') plt.show()