1.1 common distribution
| distribution | Probability or probability density | Mean E(X) | Variance D(X) |
|---|---|---|---|
| Binomial distribution | P(X=x)=Cnxpxqn−x P(X=x) = C_n^x p^x q^{n-x} P(X=x)=Cnxpxqn−x | npnpnp | np(1−p)np(1-p)np(1−p) |
| Poisson distribution | P{X=i}=e−λλii! P\lbrace X= i \rbrace = e^{-λ} \frac{λ^i}{i!} P{X=i}=e−λi!λi | λλλ | λλλ |
| Normal distribution X ~ N(μ, σ 2 σ ^ 2 σ 2) | f(x)=12πσe−(x−u)22σ2 f(x)=\frac{1}{\sqrt{2π}\sigma}e^{\frac{-(x-u)^2}{2\sigma^2}} f(x)=2πσ1e2σ2−(x−u)2 | uuu | σ2\sigma^2σ2 |
| Evenly distributed X~U[a,b] | f(x)={1b−a,a≤x≤b 0,others f(x)= \begin{cases} \frac {1} {b-a} , & a \leq x \leq b \ 0, & others \end{cases} f(x)={b−a1,a≤x≤b 0,others | a+b2\frac {a+b} {2}2a+b | (a−b)212\frac {(a-b)^2} {12}12(a−b)2 |
| Exponential distribution X~E (λ) | f(x)={λe−λx,x≥0 0,x<0 f(x)= \begin{cases} λe^{-λx} , & x \geq 0 \ 0, & x < 0 \end{cases} f(x)={λe−λx,x≥0 0,x<0 | 1λ\frac {1} {λ}λ1 | 1λ2\frac {1} {λ^2}λ21 |
| Chi square distribution | nnn | 2n2^n2n |
1.2. Other distribution
- Geometric distribution
Considering independent repeated tests, the geometric distribution describes the probability of first success after k tests, assuming that each success rate is p, P{X=n}=(1 − p)n − 1p P\lbrace X= n \rbrace = {(1-p)}^{n-1} p P{X=n}=(1 − p)n − 1P
- Negative binomial distribution
Considering independent repeated trials, the negative binomial distribution describes the probability of the trial going on until r times of success, assuming that each success rate is p, P {x = n} = cn − 1R − 1pr(1 − P) n − r P\lbrace X= n \rbrace = C_{n-1}^{r-1} p^r {(1-p)}^{n-r} P{X=n}=Cn−1r−1pr(1−p)n−r
- Hypergeometric Distribution
Hypergeometric distribution describes the sampling in a population with a total number of N, in which k elements belong to one group and the remaining n-k elements belong to another group. It is assumed that N times are extracted from the population, and the probability of containing x first group is P{X=n}=CkxCN − kn − xCNn P\lbrace X= n \rbrace = \frac {C_{k}^{x} C_{N-k}^{n-x}} {C_{N}^{n}} P{X=n}=CNnCkxCN−kn−x
- Gamma gamma distribution
It is often used to describe the distribution of waiting time for an event to occur n times in total
- Weibull distribution
Commonly used to describe the life span of a class of objects with "weakest chain" in the engineering field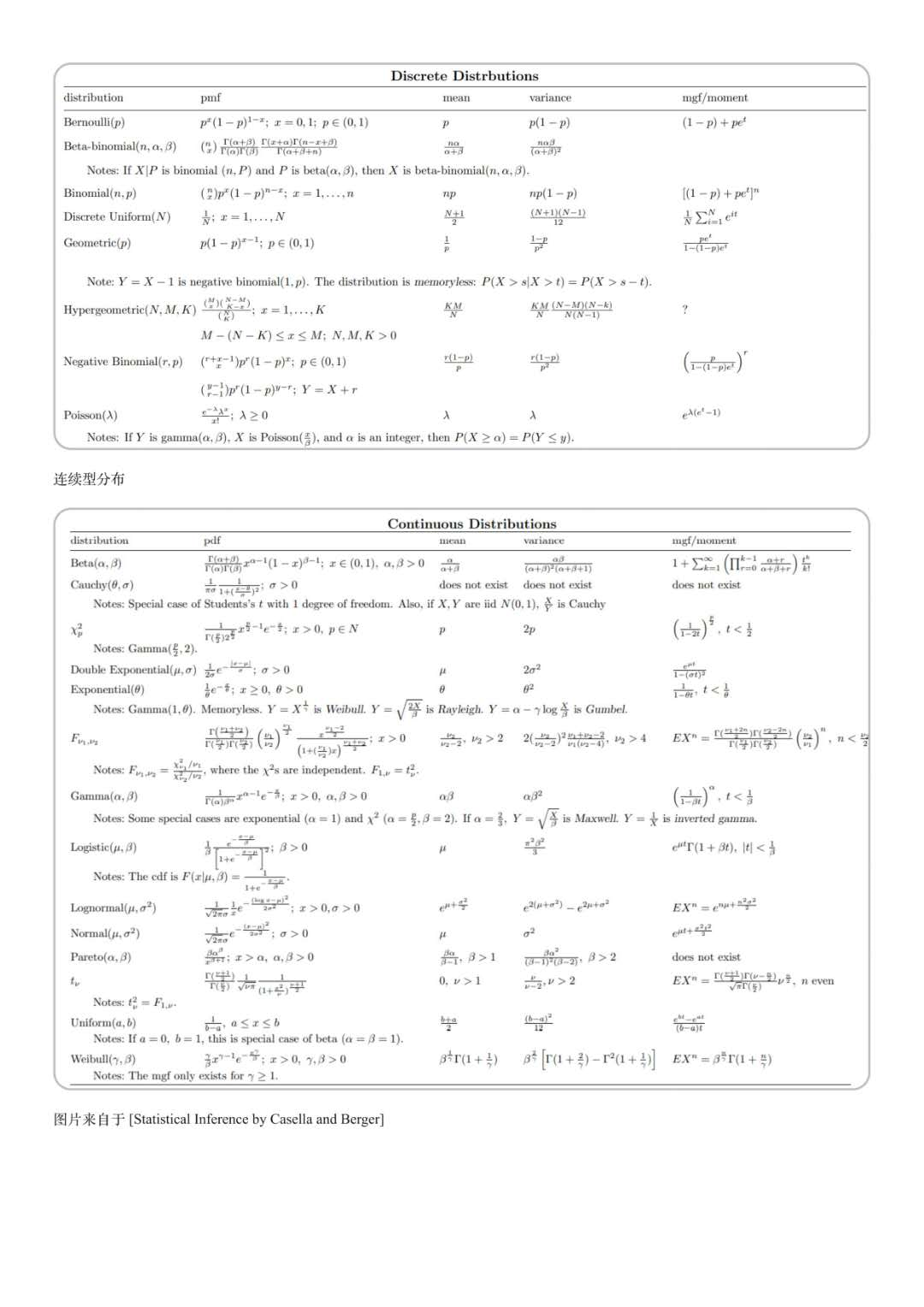
1.3.python implementation
- PDF: probability density function,
In mathematics, the probability density function of continuous random variable (which can be called density function for short if it is not confused) is a function describing the output value of the random variable and the possibility near a certain value point. - PMF: probability mass function,
In probability theory, probability mass function is the probability of discrete random variable in each specific value. - CDF: cumulative distribution
function), also known as distribution function, is the integral of probability density function, which can completely describe the probability distribution of a real random variable X.
The idea below is to use the API in the class library to achieve the desired functions
One seaborn.distplot () instructions
seaborn.distplot(a, bins=None, hist=True, kde=True, label=None) Draw the distribution map of single variable observation value flexibly. This function combines the hist function of matplotlib (automatically calculating a default bin size), kdeplot() and rugplot() functions of seaborn. It can also fit scipy.stats Distribute and plot the estimated PDF (probability distribution function) on the data. #a: Series, 1-dimensional array or list. Observe the data. If it is a series object with the name attribute, the name is used to tag the data axis. #Bins: parameter to matplotlib hist(), or None. Optional parameters. The number of histogram bins (columns). If None is filled in, the number of columns is specified by using the Freedman Diaconis rule by default. #hist: Boolean value, optional parameter. Whether to draw (standardize) histogram. #kde: Boolean value, optional parameter. Whether to draw Gauss kernel density estimation map. #label: string, optional parameter. Legend labels for the relevant components of the drawing.
Two seaborn.scatterplot () instructions
seaborn.scatterplot(x=None, y=None, hue=None, style=None, size=None, data=None, palette=None, hue_order=None, hue_norm=None, sizes=None, size_order=None, size_norm=None, markers=True, style_order=None, x_bins=None, y_bins=None, units=None, estimator=None, ci=95, n_boot=1000, alpha='auto', x_jitter=None, y_jitter=None, legend='brief', ax=None, **kwargs) //To draw a scatter chart, you can adjust parameters such as color, size, and style to display the relationship between data. #hue: variable name in data (for example, column name in 2D data): group lines that will generate different colors, which can be classification or data. #size: variable name in data (for example: column name in 2D data): group lines to be generated with different width, which can be classification or data. #style: variable name in data (for example: column name in 2D data): group variables that will be generated with different dashes or other tags. #palette: debug board name, list or dictionary type: sets the different level colors of the variables specified by hue. #hue_order: list type: Specifies the specified order in which the hue variables appear, otherwise they are determined based on the data. #hue_norm:tuple Or Normalize object #sizes:list dict Or tuple type: set the line width. When it is a number, it can also be a tuple. Specify the maximum and minimum values to be used, and other values will be normalized within the range automatically. #Units: group sampling units for variable identification. When used, a separate row will be drawn for each unit. #estimator:pandas Method name or callback function or None //Function: the method used to aggregate multiple observations of y variable at the same x level, if it isNone,All observations are drawn.
Three seaborn.lineplot () instructions
seaborn.lineplot(x=None, y=None, hue=None, size=None, style=None, data=None, palette=None, hue_order=None, hue_norm=None, sizes=None, size_order=None, size_norm=None, dashes=True, markers=None, style_order=None, units=None, estimator='mean', ci=95, n_boot=1000, sort=True, err_style='band', err_kws=None, legend='brief', ax=None, **kwargs) //To draw a line chart, you can adjust parameters such as color, size, and style to display the relationship between data. //Similar to seaborn.scatterplot
Copy the original for review
#Generate a set of random numbers that match a specific distribution //In the Numpy library, a set of random classes is provided to generate random numbers with specific distribution import numpy # Generate a set of samples with a size of 1000 that match the binomial distribution of b(10,0.5) s = numpy.random.binomial(n=10,p=0.5,size=1000) # Generate a sample set of P(1) - compliant Poisson distribution with a size of 1000 s = numpy.random.poisson(lam=1,size=1000) # In this method, the boundary value is left closed and right open s = numpy.random.uniform(low=0,high=1,size=1000) # Generate a sample set with a size of 1000 that conforms to the N(0,1) normal distribution. You can use the normal function to customize the mean value, standard deviation, or directly use standard_normal function s = numpy.random.normal(loc=0,scale=1,size=1000) s = numpy.random.standard_normal(size=1000) # Generate a sample set with a size of 1000 that conforms to the E(1/2) exponential distribution. Note that the parameter in this method is the reciprocal of the parameter λ of the exponential distribution s = numpy.random.exponential(scale=2,size=1000) #In addition to Numpy, Scipy also provides a set of methods for generating random numbers with a specific distribution # Taking the uniform distribution as an example, rvs can be used to generate the values of a set of random variables from scipy import stats stats.uniform.rvs(size=10) #PMF and PDF for statistical distribution calculation #Scipy library provides a set of methods for calculating PMF of discrete random variable and PDF of continuous random variable. from scipy import stats # Calculate PMF of binomial distribution B(10,0.5) x=range(11) p=stats.binom.pmf(x, n=10, p=0.5) # PMF calculation of Poisson distribution P(1) x=range(11) p=stats.poisson.pmf(x, mu=1) # PDF calculation of uniform distribution U(0,1) x = numpy.linspace(0,1,100) p= stats.uniform.pdf(x,loc=0, scale=1) # PDF calculation of normal distribution N(0,1) x = numpy.linspace(-3,3,1000) p= stats.norm.pdf(x,loc=0, scale=1) # PDF for calculating exponential distribution E(1) x = numpy.linspace(0,10,1000) p= stats.expon.pdf(x,loc=0,scale=1) #Calculate CDF of statistical distribution //Similar calculation probability mass/The method of density function only needs to pmf or pdf Replace with cdf,Then we can get the value of the distribution function # Taking normal distribution as an example, CDF of normal distribution N(0,1) is calculated x = numpy.linspace(-3,3,1000) p = stats.norm.cdf(x,loc=0, scale=1) #Visualization of statistical distribution #Binomial distribution #Comparing the real probability quality of binomial distribution with n=10, p=0.5 and the results of 10000 random sampling from scipy import stats import matplotlib.pyplot as plt import seaborn as sns x = range(11) # Number of successful binomial distribution (X-axis) t = stats.binom.rvs(10,0.5,size=10000) # B(10,0.5) random sampling 10000 times p = stats.binom.pmf(x, 10, 0.5) # B(10,0.5) true probability mass fig, ax = plt.subplots(1, 1) sns.distplot(t,bins=10,hist_kws={'density':True}, kde=False,label = 'Distplot from 10000 samples') sns.scatterplot(x,p,color='purple') sns.lineplot(x,p,color='purple',label='True mass density') plt.title('Binomial distribution') plt.legend(bbox_to_anchor=(1.05, 1)) #Poisson distribution #Comparing the real probability quality of Poisson distribution with the result of 10000 random sampling from scipy import stats import matplotlib.pyplot as plt import seaborn as sns x=range(11) t= stats.poisson.rvs(2,size=10000) p=stats.poisson.pmf(x, 2) fig, ax = plt.subplots(1, 1) sns.distplot(t,bins=10,hist_kws={'density':True}, kde=False,label = 'Distplot from 10000 samples') sns.scatterplot(x,p,color='purple') sns.lineplot(x,p,color='purple',label='True mass density') plt.title('Poisson distribution') plt.legend() #By comparing the probability mass functions corresponding to different parameters λ, it can be verified that as the parameters increase, the Poisson distribution becomes more and more symmetrical, and the distribution becomes more and more uniform, approaching to the normal distribution x=range(50) fig, ax = plt.subplots() for lam in [1,2,5,10,20] : p=stats.poisson.pmf(x, lam) sns.lineplot(x,p,label='lamda= '+ str(lam)) plt.title('Poisson distribution') plt.legend() #uniform distribution #Comparing the real probability density of uniform distribution of U(0,1) with the results of 10000 random sampling from scipy import stats import matplotlib.pyplot as plt import seaborn as sns x=numpy.linspace(0,1,100) t= stats.uniform.rvs(0,1,size=10000) p=stats.uniform.pdf(x, 0, 1) fig, ax = plt.subplots(1, 1) sns.distplot(t,bins=10,hist_kws={'density':True}, kde=False,label = 'Distplot from 10000 samples') sns.lineplot(x,p,color='purple',label='True mass density') plt.title('Uniforml distribution') plt.legend(bbox_to_anchor=(1.05, 1)) #Normal distribution #Comparing the real probability density of N(0,1) normal distribution with 10000 random sampling results from scipy import stats import matplotlib.pyplot as plt import seaborn as sns x=numpy.linspace(-3,3,100) t= stats.norm.rvs(0,1,size=10000) p=stats.norm.pdf(x, 0, 1) fig, ax = plt.subplots(1, 1) sns.distplot(t,bins=100,hist_kws={'density':True}, kde=False,label = 'Distplot from 10000 samples') sns.lineplot(x,p,color='purple',label='True mass density') plt.title('Normal distribution') plt.legend(bbox_to_anchor=(1.05, 1)) #Comparison of probability density functions of normal distribution with different combinations of mean and standard deviation x=numpy.linspace(-6,6,100) p=stats.norm.pdf(x, 0, 1) fig, ax = plt.subplots() for mean, std in [(0,1),(0,2),(3,1)]: p=stats.norm.pdf(x, mean, std) sns.lineplot(x,p,label='Mean: '+ str(mean) + ', std: '+ str(std)) plt.title('Normal distribution') plt.legend() #exponential distribution #Comparing the real probability density of E(1) exponential distribution with the results of 10000 random sampling from scipy import stats import matplotlib.pyplot as plt import seaborn as sns x=numpy.linspace(0,10,100) t= stats.expon.rvs(0,1,size=10000) p=stats.expon.pdf(x, 0, 1) fig, ax = plt.subplots(1, 1) sns.distplot(t,bins=100,hist_kws={'density':True}, kde=False,label = 'Distplot from 10000 samples') sns.lineplot(x,p,color='purple',label='True mass density') plt.title('Exponential distribution') plt.legend(bbox_to_anchor=(1, 1)) #Comparison of probability density functions of exponential distribution with different parameters x=numpy.linspace(0,10,100) fig, ax = plt.subplots() for scale in [0.2,0.5,1,2,5] : p=stats.expon.pdf(x, scale=scale) sns.lineplot(x,p,label='lamda= '+ str(1/scale)) plt.title('Exponential distribution') plt.legend()
2. Hypothesis test
2.1 basic concepts
Hypothesis testing is an important problem in statistical inference. When the distribution function of a population is completely unknown or only its form and parameters are unknown, in order to infer some unknown characteristics of the population, some hypotheses about the population are put forward, which are called hypothesis testing.
P value is the probability of more extreme results than the observed results when the original hypothesis is true. If the p value is very small, it means that the probability of the occurrence of the original hypothesis is very small. If it does, according to the principle of small probability, we have reason to reject the original hypothesis. The smaller the p value is, the more sufficient the reason we reject the original hypothesis. In a word, the smaller the p value is, the more significant the result is. Generally, we use P=0.05 as the critical value (one-sided test).
2.2 basic steps
A hypothesis testing problem can be divided into five steps, which must be followed no matter the details change.
- State research hypotheses, including null hypothesis and alternative hypothesis
- Collect data for validation assumptions
- Construct appropriate statistical test quantity and test
- Decide whether to accept or reject the original assumption
- Show conclusions
2.3 selection of Statistics
Choosing the right statistics is the key step of hypothesis testing. The most commonly used statistical tests include regression test, comparison test and correlation test.
- Regression test
Regression test is applicable to the case that the prediction variable is numerical type. According to the number of prediction variables and the type of result variables, it can be divided into the following categories.
| Forecast variable | Result variable | |
|---|---|---|
| Simple linear regression | Single, continuous value | Continuous value |
| Multiple linear regression | Multiple, consecutive values | Continuous value |
| Logistic regression | Continuous value | Binary category |
- Comparative test
The comparison test is applicable to the case that the forecast variable is of category type and the result variable is of numerical type. According to the grouping number of forecast variables and the number of result variables, it can be divided into the following categories.
| Forecast variable | Result variable | |
|---|---|---|
| Paired t-test | Two groups, category | Group from same population, numeric |
| ANOVA | Two or more groups, category | Single, numeric |
| MANOVA | Two or more groups, category | Two or more, numerical |
- Correlation test
Only chi square test is commonly used in association test, which is applicable to the case that both the predicted variable and the result variable are of category type.
- Nonparametric test
In addition, generally speaking, the above-mentioned parameter tests need to meet some preconditions, the samples are independent, the variance approximation and data in different groups meet the normality, so when these conditions are not satisfied, we can try to use non parameter tests instead of parameter tests.
| Nonparametric test | Parameter inspection for substitution |
|---|---|
| Spearman | Regression and correlation test |
| Sign test | T-test |
| Kruskal–Wallis | ANOVA |
| ANOSIM | MANOVA |
| Wilcoxon Rank-Sum test | Independent t-test |
| Wilcoxon Signed-rank test | Paired t-test |
2.4 two types of errors
In fact, it is possible to make mistakes in the process of hypothesis testing, and in theory, mistakes can not be completely avoided. According to the definition, errors can be divided into two types: type I error and type II error.
- A kind of mistake: reject the original hypothesis
- The second kind of mistake: accept the original assumption of mistake
A class of errors can be controlled by α value, and the α (significance level) selected in hypothesis test has a direct impact on a class of errors. α can be regarded as the greatest possibility for us to make a kind of mistake. Take 95% confidence level as an example, a=0.05, which means that the probability of rejecting a true original hypothesis is 5%. In the long run, every 20 hypothesis tests will make a mistake.
Class II errors are usually caused by small sample or high sample variance. The probability of class II errors can be expressed by β. Unlike class I errors, such errors cannot be directly controlled by setting an error rate. For the second kind of error, we can estimate it from the perspective of efficacy. First, we calculate the efficacy value 1 - β by power analysis, and then get the second kind of error estimation value β.
Generally speaking, these two kinds of errors cannot be reduced at the same time. On the premise of reducing the number of errors of the first kind, the possibility of making the number of errors of the second kind will increase. How to balance these two kinds of errors in actual cases depends on whether we are more able to accept the number of errors of the first kind or the number of errors of the second kind.
2.5 Python code practice
This section uses some examples to show how to use python for hypothesis testing.
2.5.1 normal inspection
Shapiro Wilk test is a classical normal test method.
H0: sample population obeys normal distribution
H1: the sample population does not obey the normal distribution
import numpy as np from scipy.stats import shapiro data_nonnormal = np.random.exponential(size=100) data_normal = np.random.normal(size=100) def normal_judge(data): stat, p = shapiro(data) if p > 0.05: return 'stat={:.3f}, p = {:.3f}, probably gaussian'.format(stat,p) else: return 'stat={:.3f}, p = {:.3f}, probably not gaussian'.format(stat,p) # output normal_judge(data_nonnormal) # 'stat=0.850, p = 0.000, probably not gaussian' normal_judge(data_normal) # 'stat=0.987, p = 0.415, probably gaussian'
Chi square test
Purpose: to test whether two groups of category variables are related or independent
H0: two samples are independent
H1: two groups of samples are not independent
from scipy.stats import chi2_contingency table = [[10, 20, 30],[6, 9, 17]] stat, p, dof, expected = chi2_contingency(table) print('stat=%.3f, p=%.3f' % (stat, p)) if p > 0.05: print('Probably independent') else: print('Probably dependent') # output #stat=0.272, p=0.873 #Probably independent
2.5.3 T-test
Objective: to test whether there is significant difference between the mean values of two independent sample sets
H0: mean is equal
H1: the mean value is unequal
from scipy.stats import ttest_ind import numpy as np data1 = np.random.normal(size=10) data2 = np.random.normal(size=10) stat, p = ttest_ind(data1, data2) print('stat=%.3f, p=%.3f' % (stat, p)) if p > 0.05: print('Probably the same distribution') else: print('Probably different distributions') # output # stat=-1.382, p=0.184 # Probably the same distribution
2.5.4 ANOVA
Objective: similar to t-test, ANOVA can test whether there is significant difference between the mean values of two or more independent sample sets
H0: mean is equal
H1: the mean value is unequal
from scipy.stats import f_oneway import numpy as np data1 = np.random.normal(size=10) data2 = np.random.normal(size=10) data3 = np.random.normal(size=10) stat, p = f_oneway(data1, data2, data3) print('stat=%.3f, p=%.3f' % (stat, p)) if p > 0.05: print('Probably the same distribution') else: print('Probably different distributions') # output # stat=0.189, p=0.829 # Probably the same distribution
2.5.5 Mann-Whitney U Test
Objective: to test whether the distribution of two sample sets is the same
H0: the distribution of the two sample sets is the same
H1: the distribution of the two sample sets is different
from scipy.stats import mannwhitneyu data1 = [0.873, 2.817, 0.121, -0.945, -0.055, -1.436, 0.360, -1.478, -1.637, -1.869] data2 = [1.142, -0.432, -0.938, -0.729, -0.846, -0.157, 0.500, 1.183, -1.075, -0.169] stat, p = mannwhitneyu(data1, data2) print('stat=%.3f, p=%.3f' % (stat, p)) if p > 0.05: print('Probably the same distribution') else: print('Probably different distributions') # output # stat=40.000, p=0.236 # Probably the same distribution