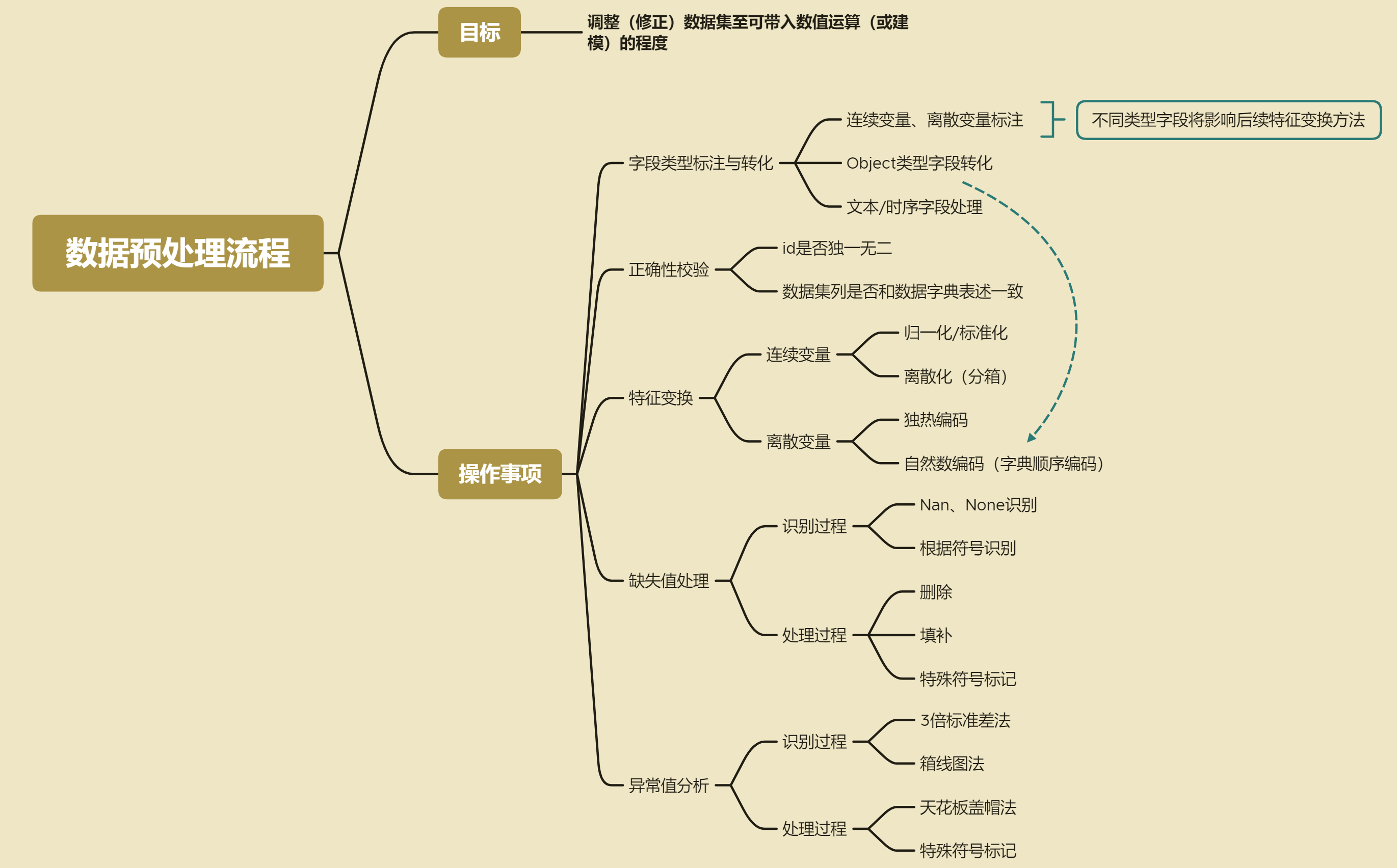
Data preprocessing process
thinking
import pandas as pd import numpy as np
data fetch
train = pd.read_csv("preprocess/train.csv")
test = pd.read_csv("preprocess/test.csv")
Stochastic forest model prediction
Feature selection – Pearson correlation coefficient
(train.shape, test.shape)
((201917, 1700), (123623, 1699))
# Extract feature name
features = train.columns.tolist()
features.remove('card_id')
features.remove("target")
featureSelect = features
# Calculate correlation coefficient
corr = []
for fea in featureSelect:
corr.append(abs(train[[fea,'target']].fillna(0).corr().values[0][1]))
# Take the features of top300 for modeling, and the specific quantity can be selected
se = pd.Series(corr, index=featureSelect).sort_values(ascending=False)
feature_select = ['card_id'] + se[:300].index.tolist()
# Output results
train = train[feature_select + ['target']]
test = test[feature_select]
Grid search parameters
from sklearn.metrics import mean_squared_error from sklearn.ensemble import RandomForestRegressor from sklearn.model_selection import GridSearchCV
| Name | Description |
|---|---|
| criterion | Rule evaluation index or loss function, default Gini coefficient, optional information entropy |
| splitter | Tree model growth mode: by default, it grows in the fastest way to reduce the value of loss function. It can be divided randomly according to certain conditions |
| max_depth | The maximum growth depth of the tree, similar to max_iter, that is, how many iterations in total |
| min_samples_split | Minimum number of samples required for internal node re Division |
| min_samples_leaf | The leaf node contains the minimum number of samples |
| min_weight_fraction_leaf | Minimum weight sum required for leaf nodes |
| max_features | How many features can be brought in during segmentation to select the segmentation rules |
| random_state | Random number seed |
| max_leaf_nodes | Maximum number of leaf nodes |
| min_impurity_decrease | At least the loss value that needs to be reduced for data set re division |
| min_impurity_split | The minimum impurity required for data set repartition will be removed in version 0.25 |
| class_weight | Weight of various samples |
then the selection of grid search tools. With the continuous improvement of sklearn, there are more and more grid search tools to choose from, but on the whole, it is actually a trade-off between efficiency and accuracy. Some grid search tools have slow execution efficiency but guaranteed result accuracy due to global enumeration (such as GridSearchCV). If you are willing to sacrifice accuracy for execution efficiency, there are also many tools to choose from, Such as RandomizedSearchCV. Of course, in the latest version of sklearn, there is also a more efficient search strategy - HalvingGridSearchCV. This method first compares two by two, and then filters the parameters layer by layer. It supports both HalvingGridSearchCV and HalvingRandomSearchCV. Note that this function is only supported in the latest version of sklearn, that is, version 0.24. The emergence of this function is also one of the biggest changes in version 0.24. The addition of this function will further reduce the computing resources required for grid search and speed up grid search.
based on the data of this competition, in the actual process of grid search, it is recommended to use randomized searchcv to determine the approximate range, and then use GridSearchCV to search for specific parameter values with high precision. Of course, if you are using the latest version of sklearn, you can also consider using the Halving method for search. In the course of public lecture, due to the limited time, we set a relatively small parameter space to search on the premise of roughly determining the optimal parameter range:
features = train.columns.tolist()
features.remove("card_id")
features.remove("target")
parameter_space = {
"n_estimators": [79, 80, 81],
"min_samples_leaf": [29, 30, 31],
"min_samples_split": [2, 3],
"max_depth": [9, 10],
"max_features": ["auto", 80]
}
clf = RandomForestRegressor(criterion="mse",
n_jobs=64,
random_state=22)
# train[feature].isnull().sum
grid = GridSearchCV(clf, parameter_space, cv=2, scoring="neg_mean_squared_error") grid.fit(train[features].values, train['target'].values)
GridSearchCV(cv=2, estimator=RandomForestRegressor(n_jobs=64, random_state=22),
param_grid={'max_depth': [9, 10], 'max_features': ['auto', 80],
'min_samples_leaf': [29, 30, 31],
'min_samples_split': [2, 3],
'n_estimators': [79, 80, 81]},
scoring='neg_mean_squared_error')
grid.best_estimator_
RandomForestRegressor(max_depth=10, max_features=80, min_samples_leaf=31,
n_estimators=80, n_jobs=64, random_state=22)