1 Introduction
A data classification model based on genetic algorithm and random forest.
1. The basic principle of random forest random forest was proposed by Leo Breiman(2001). It uses bootstrap resampling technology to repeatedly randomly select # b samples from the original training sample set # N # to generate a new training self-help sample set, and then generate # b classification trees according to the self-help sample set to form a random forest, The classification result of new data depends on the score formed by the number of votes in the classification tree. The main feature of random forest is that it will not produce over fitting phenomenon when processing high-dimensional data. It can give the importance score of variables while classifying. According to the score, the variables that play an important role in classification can be screened out.
2. Basic principle of genetic algorithm genetic algorithm was proposed by J. Holland of Michigan University in 1975. It is a random search algorithm based on the natural selection and genetic mechanism of organisms. Its basic principle is evolutionary mechanism and natural selection law. The characteristic of genetic algorithm is to use simple coding technology to represent complex structure, generate alternative solution set through genetic operation of coding - reproduction, crossover and mutation, and conduct directional search through the selection mechanism of survival of the fittest. Evolutionary algorithm does not need to understand all the characteristics of the problem, but can solve the problem through the evolutionary process reflecting the evolutionary mechanism.
Garf ¢ uses the ¢ RF ¢ model to evaluate the role of variables in sample classification, and uses the ¢ permutation ¢ method to determine the boundary value of feature screening as the basis for the final determination of feature variables. In order to reduce the interference of noise variables on the evaluation results of ^ RF ^ variables, each ^ RF ^ model only contains some variables selected by ^ GA ^ algorithm, and variable screening steps are added in the genetic process to further reduce the impact of noise variables. The heuristic characteristic of GA algorithm makes the variables with strong classification have more opportunities to be evaluated, so as to enhance the stability of GA evaluation results; At the same time, the "variation" process of GA # algorithm makes the search results converge in a certain direction and have strong diversity, so that the variables with weak classification can also get a certain opportunity to be evaluated. The above two characteristics of GA # algorithm not only ensure the depth of variable evaluation, but also take into account the breadth of evaluation. In the variable screening within the genetic process, the "permutation" method is used to obtain the empirical distribution of the importance score of no difference variables between groups, and the boundary value of variable screening is determined adaptively according to the empirical distribution.
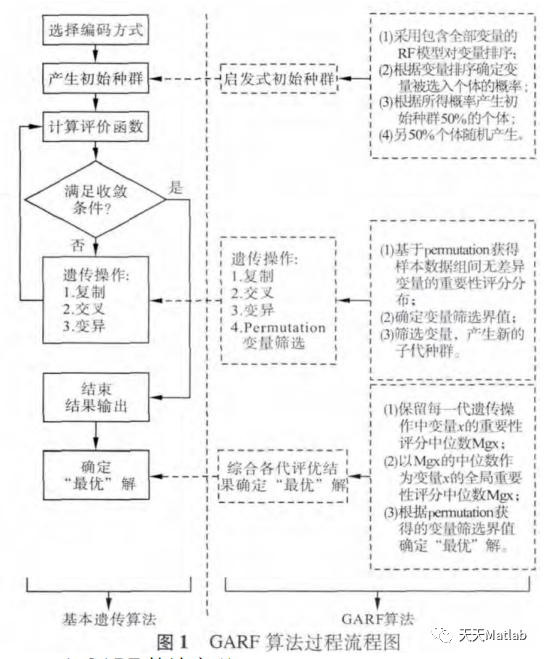
Part 2 code
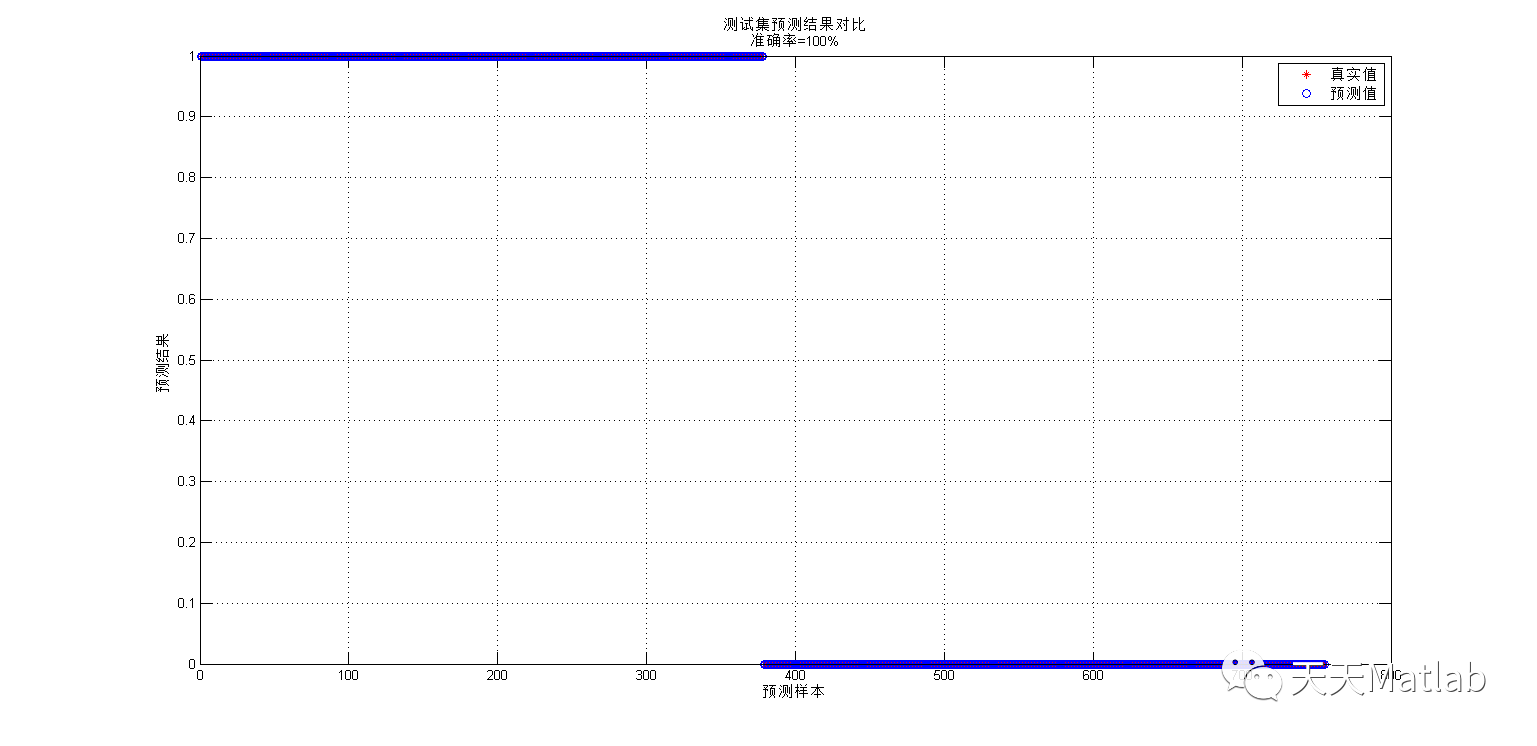
%**************************************************************eb/packages/randomForest/randomForest.pdf%**************************************************************%function [Y_hat votes] = classRF_predict(X,model, extra_options)% requires 2 arguments% X: data matrix% model: generated via classRF_train function% extra_options.predict_all = predict_all if set will send all the prediction. %%% Returns% Y_hat - prediction for the data% votes - unnormalized weights for the model% prediction_per_tree - per tree prediction. the returned object .% If predict.all=TRUE, then the individual component of the returned object is a character% matrix where each column contains the predicted class by a tree in the forest.%%% Not yet implemented% proximityfunction [Y_new, votes, prediction_per_tree] = classRF_predict(X,model, extra_options) if nargin<2 error('need atleast 2 parameters,X matrix and model'); end if exist('extra_options','var') if isfield(extra_options,'predict_all') predict_all = extra_options.predict_all; end end if ~exist('predict_all','var'); predict_all=0;end [Y_hat,prediction_per_tree,votes] = mexClassRF_predict(X',model.nrnodes,model.ntree,model.xbestsplit,model.classwt,model.cutoff,model.treemap,model.nodestatus,model.nodeclass,model.bestvar,model.ndbigtree,model.nclass, predict_all); %keyboard votes = votes'; clear mexClassRF_predict Y_new = double(Y_hat); new_labels = model.new_labels; orig_labels = model.orig_labels; for i=1:length(orig_labels) Y_new(find(Y_hat==new_labels(i)))=Inf; Y_new(isinf(Y_new))=orig_labels(i); end 1; 3 simulation results
4 references
[1] Zhang Zhongnan, Luo Weizhen A EEG signal classification model based on genetic algorithm and random forest: cn108615024a [P] two thousand and eighteen
Blogger profile: good at matlab simulation in intelligent optimization algorithm, neural network prediction, signal processing, cellular automata, image processing, path planning, UAV and other fields. Relevant matlab code problems can be exchanged through private letters.
Some theories cite online literature. If there is infringement, contact the blogger and delete it.