This article aims to gradually get familiar with the implementation of the overall architecture of hudi through a certain function, and will not discuss the implementation details of the algorithm
hudi newcomer, if you have any questions, please correct them
spark : version, 3.1.2
hudi : branch, master
Time: 2022/02/06 First Edition
Objective: to reduce the amount of data in data scan by changing the way of data layout.
Take a simple Chestnut:
- A text table contains two fields: id and name
There are two data files, a.parquet and b.parquet
- a.parquet data 2,zs, 1,ls, 4, Wu, 3, TS
- b.parquet data 1,ls, 2,zs, 4, Wu, 5, TS
- At this time, we need to make statistics on the number of filters for id = 2. We need to scan a.parquet and b.parquet files
After sorting the data and making Min/Max index records
- a.parquet data 1,ls, 1,ls, 2,zs, 2,zs min ID: 1 | Max ID: 2
- b.parquet data 3,ts, 4,wu, 4,wu, 5, TS min ID: 3 | Max ID: 5
- At this time, we need to make statistics on the number of filters for id = 2. We only need to scan a.parquet file
Query phase
Entry class, DefaultSource, createRelation method, create Relation
type condition:
- hoodie.datasource.query.type is read_ When optimized, Hoodie table. Type is cow or mor
- hoodie. datasource. query. When the type is snapshot, Hoodie table. Type is cow
After the above conditions are met, Hadoop fsrelation will be created. The HoodieFileIndex needs to be created and initialized. When initializing the HoodieFileIndex, refresh0() will be called to build the fileSice to be queried. If it is a partition table, the file paths and related information under all partition directories of the list will still be read and cached in cachedialinputfileslices
private def refresh0(): Unit = {
val startTime = System.currentTimeMillis()
// Load file s for all partitions
val partitionFiles = loadPartitionPathFiles()
val allFiles = partitionFiles.values.reduceOption(_ ++ _)
.getOrElse(Array.empty[FileStatus])
metaClient.reloadActiveTimeline()
val activeInstants = metaClient.getActiveTimeline.getCommitsTimeline.filterCompletedInstants
val latestInstant = activeInstants.lastInstant()
fileSystemView = new HoodieTableFileSystemView(metaClient, activeInstants, allFiles)
val queryInstant = if (specifiedQueryInstant.isDefined) {
specifiedQueryInstant
} else if (latestInstant.isPresent) {
Some(latestInstant.get.getTimestamp)
} else {
None
}
(tableType, queryType) match {
case (MERGE_ON_READ, QUERY_TYPE_SNAPSHOT_OPT_VAL) =>
// Fetch and store latest base and log files, and their sizes
// All FileSlices will be stored here
cachedAllInputFileSlices = partitionFiles.map(p => {
val latestSlices = if (latestInstant.isPresent) {
fileSystemView.getLatestMergedFileSlicesBeforeOrOn(p._1.partitionPath, queryInstant.get)
.iterator().asScala.toSeq
} else {
Seq()
}
(p._1, latestSlices)
})
cachedFileSize = cachedAllInputFileSlices.values.flatten.map(fileSlice => {
if (fileSlice.getBaseFile.isPresent) {
fileSlice.getBaseFile.get().getFileLen + fileSlice.getLogFiles.iterator().asScala.map(_.getFileSize).sum
} else {
fileSlice.getLogFiles.iterator().asScala.map(_.getFileSize).sum
}
}).sum
case (_, _) =>
// Fetch and store latest base files and its sizes
cachedAllInputFileSlices = partitionFiles.map(p => {
val fileSlices = specifiedQueryInstant
.map(instant =>
fileSystemView.getLatestFileSlicesBeforeOrOn(p._1.partitionPath, instant, true))
.getOrElse(fileSystemView.getLatestFileSlices(p._1.partitionPath))
.iterator().asScala.toSeq
(p._1, fileSlices)
})
cachedFileSize = cachedAllInputFileSlices.values.flatten.map(fileSliceSize).sum
}Houdiefileindex inherits org apache. spark. sql. execution. datasources. Fileindex implements the listFiles interface, which is used to read the zIndex index and achieve the effect of partition filtering. Houdiefileindex is used as the implementation of HadoopFsRelation's location and as the data of data input
listFiles implementation of houdiefileindex
Lookupcandidate files inzindex will use ZIndex to find the file to be read
// code snippet
override def listFiles(partitionFilters: Seq[Expression],
dataFilters: Seq[Expression]): Seq[PartitionDirectory] = {
// Look up candidate files names in the Z-index, if all of the following conditions are true
// - Data-skipping is enabled
// - Z-index is present
// - List of predicates (filters) is present
val candidateFilesNamesOpt: Option[Set[String]] =
lookupCandidateFilesInZIndex(dataFilters) match {
case Success(opt) => opt
case Failure(e) =>
if (e.isInstanceOf[AnalysisException]) {
logDebug("Failed to relay provided data filters to Z-index lookup", e)
} else {
logError("Failed to lookup candidate files in Z-index", e)
}
Option.empty
}
...
}Look first File data of zindex index / zindex/20220202225600359/part-00000-3b9cdcd9-28ed-4cef-8f97-2bb6097b1445-c000.snappy.parquet (the example here is zorder sorting by name and id). It can be seen that each row of data corresponds to a data file, and the maximum and minimum values of column name and column id of each data file are recorded to realize data filtering.
(Note: the data is test data and may not conform to the filtering logic, because each data file has only one line of data and does not meet the condition of compact)
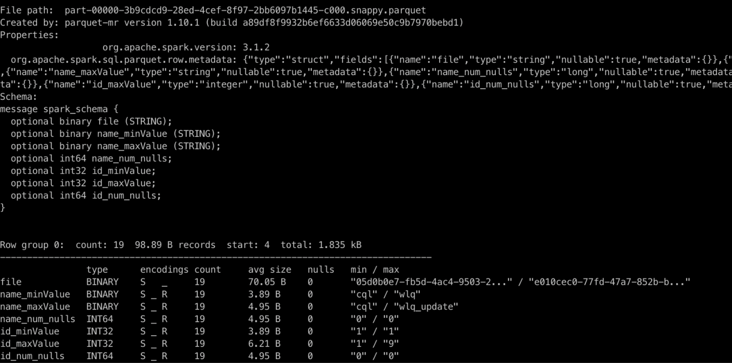
{"file": "859ccf12-253f-40d5-ba0b-a831e48e4f16-0_0-45-468_20220202155755496.parquet", "name_minValue": "cql", "name_maxValue": "wlq", "name_num_nulls": 0, "id_minValue": 1, "id_maxValue": 9, "id_num_nulls": 0}
{"file": "9f3054c8-5f57-45c8-8bdf-400707edd2d3-0_0-26-29_20220202223215267.parquet", "name_minValue": "cql", "name_maxValue": "wlq_update", "name_num_nulls": 0, "id_minValue": 1, "id_maxValue": 1, "id_num_nulls": 0}
{"file": "9c76a87b-8263-41c5-a830-08698b27ec0f-0_0-49-440_20220202160249241.parquet", "name_minValue": "cql", "name_maxValue": "cql", "name_num_nulls": 0, "id_minValue": 1, "id_maxValue": 1, "id_num_nulls": 0}lookupCandidateFilesInZIndex implementation
private def lookupCandidateFilesInZIndex(queryFilters: Seq[Expression]): Try[Option[Set[String]]] = Try {
// . zindex path, in tablename / hoodie/. zindex
val indexPath = metaClient.getZindexPath
val fs = metaClient.getFs
if (!enableDataSkipping() || !fs.exists(new Path(indexPath)) || queryFilters.isEmpty) {
// scalastyle:off return
return Success(Option.empty)
// scalastyle:on return
}
// Collect all index tables present in `.zindex` folder
val candidateIndexTables =
fs.listStatus(new Path(indexPath))
.filter(_.isDirectory)
.map(_.getPath.getName)
.filter(f => completedCommits.contains(f))
.sortBy(x => x)
if (candidateIndexTables.isEmpty) {
// scalastyle:off return
return Success(Option.empty)
// scalastyle:on return
}
val dataFrameOpt = try {
Some(spark.read.load(new Path(indexPath, candidateIndexTables.last).toString))
} catch {
case t: Throwable =>
logError("Failed to read Z-index; skipping", t)
None
}
dataFrameOpt.map(df => {
val indexSchema = df.schema
// The filter expression is constructed through the schema of the index file and the pushed queryscheme
val indexFilter =
queryFilters.map(createZIndexLookupFilter(_, indexSchema))
.reduce(And)
logInfo(s"Index filter condition: $indexFilter")
df.persist()
// Get all file s
val allIndexedFileNames =
df.select("file")
.collect()
.map(_.getString(0))
.toSet
// Filter out the file s that meet the conditions
val prunedCandidateFileNames =
df.where(new Column(indexFilter))
.select("file")
.collect()
.map(_.getString(0))
.toSet
df.unpersist()
// NOTE: Z-index isn't guaranteed to have complete set of statistics for every
// base-file: since it's bound to clustering, which could occur asynchronously
// at arbitrary point in time, and is not likely to touching all of the base files.
//
// To close that gap, we manually compute the difference b/w all indexed (Z-index)
// files and all outstanding base-files, and make sure that all base files not
// represented w/in Z-index are included in the output of this method
// Not all historical files or partitions use the zorder index, so the file pushed down may not be all the query results. For example, there is a historical file a.parquet. At this time, zorder optimization is added without brushing the history. The data files b.parquet and c.parquet are added. At this time, the zindex index only has the information of b.parquet, not a.parquet, If used directly, the data will be. Therefore, if zindex hits b.parquet, you only need to exclude c.parquet. Use the cachedialinputfileslices - c.parquet above to query the a.parquet + b.parquet file and filter out c.parqeut
val notIndexedFileNames =
lookupFileNamesMissingFromIndex(allIndexedFileNames)
prunedCandidateFileNames ++ notIndexedFileNames
})
}Generation phase
// Execute after each write hoodie.clustering.inline = 'true' hoodie.clustering.inline.max.commits = '1' hoodie.layout.optimize.strategy = 'z-order' hoodie.layout.optimize.enable = 'true' hoodie.clustering.plan.strategy.sort.columns = 'name,id'
Class abstracthoudiewriteclient will run tableservicesinline to perform relevant compact and other operations, including the generation process of zindex, after commitStats releases the lock. The process is mainly divided into two steps: first, sorting, and then generating zindex file according to replace
Note: the above sorting will not occur in the core process of spark tasks (and can be executed asynchronously), and will not affect the commit submission of the next spark data write
In the runTableServicesInline method, judge the inline_ Is the cluster enabled
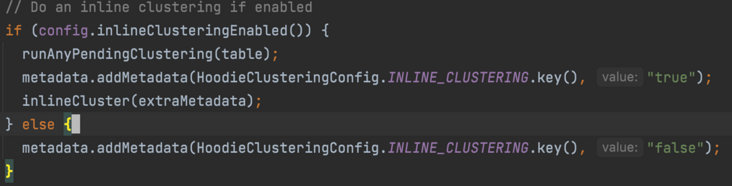
Focus on the inlineCluster method, and finally use the cluster implemented by the subclass SparkRDDWriteClient
@Override
public HoodieWriteMetadata<JavaRDD<WriteStatus>> cluster(String clusteringInstant, boolean shouldComplete) {
HoodieSparkTable<T> table = HoodieSparkTable.create(config, context, config.isMetadataTableEnabled());
preWrite(clusteringInstant, WriteOperationType.CLUSTER, table.getMetaClient());
HoodieTimeline pendingClusteringTimeline = table.getActiveTimeline().filterPendingReplaceTimeline();
HoodieInstant inflightInstant = HoodieTimeline.getReplaceCommitInflightInstant(clusteringInstant);
if (pendingClusteringTimeline.containsInstant(inflightInstant)) {
rollbackInflightClustering(inflightInstant, table);
table.getMetaClient().reloadActiveTimeline();
}
clusteringTimer = metrics.getClusteringCtx();
LOG.info("Starting clustering at " + clusteringInstant);
// Build a Partitioner as needed to sort the data, and then return the metadata
HoodieWriteMetadata<JavaRDD<WriteStatus>> clusteringMetadata = table.cluster(context, clusteringInstant);
JavaRDD<WriteStatus> statuses = clusteringMetadata.getWriteStatuses();
// TODO : Where is shouldComplete used ?
if (shouldComplete && clusteringMetadata.getCommitMetadata().isPresent()) {
// Generate zindex
completeTableService(TableServiceType.CLUSTER, clusteringMetadata.getCommitMetadata().get(), statuses, table, clusteringInstant);
}
return clusteringMetadata;
}Sorting stage
table. The cluster will eventually create a SparkExecuteClusteringCommitActionExecutor to perform related operations
@Override
public HoodieWriteMetadata<JavaRDD<WriteStatus>> execute() {
HoodieInstant instant = HoodieTimeline.getReplaceCommitRequestedInstant(instantTime);
// Mark instant as clustering inflight
table.getActiveTimeline().transitionReplaceRequestedToInflight(instant, Option.empty());
table.getMetaClient().reloadActiveTimeline();
final Schema schema = HoodieAvroUtils.addMetadataFields(new Schema.Parser().parse(config.getSchema()));
// Use the function SparkSortAndSizeExecutionStrategy for sorting optimization and performClustering
HoodieWriteMetadata<JavaRDD<WriteStatus>> writeMetadata = ((ClusteringExecutionStrategy<T, JavaRDD<HoodieRecord<? extends HoodieRecordPayload>>, JavaRDD<HoodieKey>, JavaRDD<WriteStatus>>)
ReflectionUtils.loadClass(config.getClusteringExecutionStrategyClass(),
new Class<?>[] {HoodieTable.class, HoodieEngineContext.class, HoodieWriteConfig.class}, table, context, config))
.performClustering(clusteringPlan, schema, instantTime);
JavaRDD<WriteStatus> writeStatusRDD = writeMetadata.getWriteStatuses();
JavaRDD<WriteStatus> statuses = updateIndex(writeStatusRDD, writeMetadata);
writeMetadata.setWriteStats(statuses.map(WriteStatus::getStat).collect());
writeMetadata.setPartitionToReplaceFileIds(getPartitionToReplacedFileIds(writeMetadata));
commitOnAutoCommit(writeMetadata);
if (!writeMetadata.getCommitMetadata().isPresent()) {
HoodieCommitMetadata commitMetadata = CommitUtils.buildMetadata(writeMetadata.getWriteStats().get(), writeMetadata.getPartitionToReplaceFileIds(),
extraMetadata, operationType, getSchemaToStoreInCommit(), getCommitActionType());
writeMetadata.setCommitMetadata(Option.of(commitMetadata));
}
return writeMetadata;
}Performclusteringwithrecordsdd implementation of SparkSortAndSizeExecutionStrategy
@Override
public JavaRDD<WriteStatus> performClusteringWithRecordsRDD(final JavaRDD<HoodieRecord<T>> inputRecords, final int numOutputGroups,
final String instantTime, final Map<String, String> strategyParams, final Schema schema,
final List<HoodieFileGroupId> fileGroupIdList, final boolean preserveHoodieMetadata) {
LOG.info("Starting clustering for a group, parallelism:" + numOutputGroups + " commit:" + instantTime);
Properties props = getWriteConfig().getProps();
props.put(HoodieWriteConfig.BULKINSERT_PARALLELISM_VALUE.key(), String.valueOf(numOutputGroups));
// We are calling another action executor - disable auto commit. Strategy is only expected to write data in new files.
props.put(HoodieWriteConfig.AUTO_COMMIT_ENABLE.key(), Boolean.FALSE.toString());
props.put(HoodieStorageConfig.PARQUET_MAX_FILE_SIZE.key(), String.valueOf(getWriteConfig().getClusteringTargetFileMaxBytes()));
HoodieWriteConfig newConfig = HoodieWriteConfig.newBuilder().withProps(props).build();
// Here, focus on the getPartitioner method. If you enable Hoodie layout. optimize. Enable, the rddspatialcurveoptimizesortpartitioner will be returned, and finally the repartitionRecords of the partitioner will be called
return (JavaRDD<WriteStatus>) SparkBulkInsertHelper.newInstance().bulkInsert(inputRecords, instantTime, getHoodieTable(), newConfig,
false, getPartitioner(strategyParams, schema), true, numOutputGroups, new CreateHandleFactory(preserveHoodieMetadata));
}The repartitionRecords of RDDSpatialCurveOptimizationSortPartitioner will be based on Hoodie layout. optimize. curve. build. Method calls the createOptimizedDataFrameByXXX method of OrderingIndexHelper
public static Dataset<Row> createOptimizedDataFrameByMapValue(Dataset<Row> df, List<String> sortCols, int fileNum, String sortMode) {
Map<String, StructField> columnsMap = Arrays.stream(df.schema().fields()).collect(Collectors.toMap(e -> e.name(), e -> e));
int fieldNum = df.schema().fields().length;
List<String> checkCols = sortCols.stream().filter(f -> columnsMap.containsKey(f)).collect(Collectors.toList());
if (sortCols.size() != checkCols.size()) {
return df;
}
// only one col to sort, no need to use z-order
if (sortCols.size() == 1) {
return df.repartitionByRange(fieldNum, org.apache.spark.sql.functions.col(sortCols.get(0)));
}
Map<Integer, StructField> fieldMap = sortCols
.stream().collect(Collectors.toMap(e -> Arrays.asList(df.schema().fields()).indexOf(columnsMap.get(e)), e -> columnsMap.get(e)));
// do optimize
// It can be seen that two sort curves are currently supported, z and hilbert
JavaRDD<Row> sortedRDD = null;
switch (HoodieClusteringConfig.BuildLayoutOptimizationStrategy.fromValue(sortMode)) {
case ZORDER:
sortedRDD = createZCurveSortedRDD(df.toJavaRDD(), fieldMap, fieldNum, fileNum);
break;
case HILBERT:
sortedRDD = createHilbertSortedRDD(df.toJavaRDD(), fieldMap, fieldNum, fileNum);
break;
default:
throw new IllegalArgumentException(String.format("new only support z-order/hilbert optimize but find: %s", sortMode));
}
// create new StructType
List<StructField> newFields = new ArrayList<>();
newFields.addAll(Arrays.asList(df.schema().fields()));
newFields.add(new StructField("Index", BinaryType$.MODULE$, true, Metadata.empty()));
// create new DataFrame
return df.sparkSession().createDataFrame(sortedRDD, StructType$.MODULE$.apply(newFields)).drop("Index");
}Generate Index stage
Go back to the cluster method of SparkRDDWriteClient, and finally call completeableservice to perform the commit operation
private void completeClustering(HoodieReplaceCommitMetadata metadata, JavaRDD<WriteStatus> writeStatuses,
HoodieTable<T, JavaRDD<HoodieRecord<T>>, JavaRDD<HoodieKey>, JavaRDD<WriteStatus>> table,
String clusteringCommitTime) {
List<HoodieWriteStat> writeStats = metadata.getPartitionToWriteStats().entrySet().stream().flatMap(e ->
e.getValue().stream()).collect(Collectors.toList());
if (writeStats.stream().mapToLong(s -> s.getTotalWriteErrors()).sum() > 0) {
throw new HoodieClusteringException("Clustering failed to write to files:"
+ writeStats.stream().filter(s -> s.getTotalWriteErrors() > 0L).map(s -> s.getFileId()).collect(Collectors.joining(",")));
}
try {
HoodieInstant clusteringInstant = new HoodieInstant(HoodieInstant.State.INFLIGHT, HoodieTimeline.REPLACE_COMMIT_ACTION, clusteringCommitTime);
this.txnManager.beginTransaction(Option.of(clusteringInstant), Option.empty());
finalizeWrite(table, clusteringCommitTime, writeStats);
writeTableMetadataForTableServices(table, metadata,clusteringInstant);
// Update outstanding metadata indexes
if (config.isLayoutOptimizationEnabled()
&& !config.getClusteringSortColumns().isEmpty()) {
// Update metadata index
table.updateMetadataIndexes(context, writeStats, clusteringCommitTime);
}
LOG.info("Committing Clustering " + clusteringCommitTime + ". Finished with result " + metadata);
table.getActiveTimeline().transitionReplaceInflightToComplete(
HoodieTimeline.getReplaceCommitInflightInstant(clusteringCommitTime),
Option.of(metadata.toJsonString().getBytes(StandardCharsets.UTF_8)));
} catch (Exception e) {
throw new HoodieClusteringException("unable to transition clustering inflight to complete: " + clusteringCommitTime, e);
} finally {
this.txnManager.endTransaction();
}
WriteMarkersFactory.get(config.getMarkersType(), table, clusteringCommitTime)
.quietDeleteMarkerDir(context, config.getMarkersDeleteParallelism());
if (clusteringTimer != null) {
long durationInMs = metrics.getDurationInMs(clusteringTimer.stop());
try {
metrics.updateCommitMetrics(HoodieActiveTimeline.parseDateFromInstantTime(clusteringCommitTime).getTime(),
durationInMs, metadata, HoodieActiveTimeline.REPLACE_COMMIT_ACTION);
} catch (ParseException e) {
throw new HoodieCommitException("Commit time is not of valid format. Failed to commit compaction "
+ config.getBasePath() + " at time " + clusteringCommitTime, e);
}
}
LOG.info("Clustering successfully on commit " + clusteringCommitTime);
}updateZIndex method of houdiesparkcopyonwritetable
private void updateZIndex(
@Nonnull HoodieEngineContext context,
@Nonnull List<HoodieWriteStat> updatedFilesStats,
@Nonnull String instantTime
) throws Exception {
String sortColsList = config.getClusteringSortColumns();
String basePath = metaClient.getBasePath();
String indexPath = metaClient.getZindexPath();
// Get the instant time of all commit and replaceCommit
List<String> completedCommits =
metaClient.getCommitsTimeline()
.filterCompletedInstants()
.getInstants()
.map(HoodieInstant::getTimestamp)
.collect(Collectors.toList());
// Gets the path of the newly written data file
List<String> touchedFiles =
updatedFilesStats.stream()
.map(s -> new Path(basePath, s.getPath()).toString())
.collect(Collectors.toList());
if (touchedFiles.isEmpty() || StringUtils.isNullOrEmpty(sortColsList) || StringUtils.isNullOrEmpty(indexPath)) {
return;
}
LOG.info(String.format("Updating Z-index table (%s)", indexPath));
List<String> sortCols = Arrays.stream(sortColsList.split(","))
.map(String::trim)
.collect(Collectors.toList());
HoodieSparkEngineContext sparkEngineContext = (HoodieSparkEngineContext)context;
// Fetch table schema to appropriately construct Z-index schema
Schema tableWriteSchema =
HoodieAvroUtils.createHoodieWriteSchema(
new TableSchemaResolver(metaClient).getTableAvroSchemaWithoutMetadataFields()
);
// Update index file
ZOrderingIndexHelper.updateZIndexFor(
sparkEngineContext.getSqlContext().sparkSession(),
AvroConversionUtils.convertAvroSchemaToStructType(tableWriteSchema),
touchedFiles,
sortCols,
indexPath,
instantTime,
completedCommits
);
LOG.info(String.format("Successfully updated Z-index at instant (%s)", instantTime));
}ZOrderingIndexHelper.updateZIndexFor
public static void updateZIndexFor(
@Nonnull SparkSession sparkSession,
@Nonnull StructType sourceTableSchema,
@Nonnull List<String> sourceBaseFiles,
@Nonnull List<String> zorderedCols,
@Nonnull String zindexFolderPath,
@Nonnull String commitTime,
@Nonnull List<String> completedCommits
) {
FileSystem fs = FSUtils.getFs(zindexFolderPath, sparkSession.sparkContext().hadoopConfiguration());
// Compose new Z-index table for the given source base files
// Read the metadata information newly written to the data file and use it to build the index
Dataset<Row> newZIndexDf =
buildZIndexTableFor(
sparkSession,
sourceBaseFiles,
zorderedCols.stream()
.map(col -> sourceTableSchema.fields()[sourceTableSchema.fieldIndex(col)])
.collect(Collectors.toList())
);
try {
//
// Z-Index has the following folder structure:
//
// .hoodie/
// ├── .zindex/
// │ ├── <instant>/
// │ │ ├── <part-...>.parquet
// │ │ └── ...
//
// If index is currently empty (no persisted tables), we simply create one
// using clustering operation's commit instance as it's name
Path newIndexTablePath = new Path(zindexFolderPath, commitTime);
// If zindex does not exist originally. If it is written for the first time, it will be written directly. Otherwise, the merge history will be appended
if (!fs.exists(new Path(zindexFolderPath))) {
newZIndexDf.repartition(1)
.write()
.format("parquet")
.mode("overwrite")
.save(newIndexTablePath.toString());
return;
}
// Filter in all index tables (w/in {@code .zindex} folder)
// obtain. All instant folders under zindex directory
List<String> allIndexTables =
Arrays.stream(
fs.listStatus(new Path(zindexFolderPath))
)
.filter(FileStatus::isDirectory)
.map(f -> f.getPath().getName())
.collect(Collectors.toList());
// Compile list of valid index tables that were produced as part
// of previously successfully committed iterations
List<String> validIndexTables =
allIndexTables.stream()
.filter(completedCommits::contains)
.sorted()
.collect(Collectors.toList());
List<String> tablesToCleanup =
allIndexTables.stream()
.filter(f -> !completedCommits.contains(f))
.collect(Collectors.toList());
Dataset<Row> finalZIndexDf;
// Before writing out new version of the Z-index table we need to merge it
// with the most recent one that were successfully persisted previously
if (validIndexTables.isEmpty()) {
finalZIndexDf = newZIndexDf;
} else {
// NOTE: That Parquet schema might deviate from the original table schema (for ex,
// by upcasting "short" to "integer" types, etc), and hence we need to re-adjust it
// prior to merging, since merging might fail otherwise due to schemas incompatibility
finalZIndexDf =
tryMergeMostRecentIndexTableInto(
sparkSession,
newZIndexDf,
// Load current most recent Z-index table
sparkSession.read().load(
new Path(zindexFolderPath, validIndexTables.get(validIndexTables.size() - 1)).toString()
)
);
// Clean up all index tables (after creation of the new index)
tablesToCleanup.addAll(validIndexTables);
}
// Persist new Z-index table
finalZIndexDf
.repartition(1)
.write()
.format("parquet")
.save(newIndexTablePath.toString());
// Clean up residual Z-index tables that have might have been dangling since
// previous iterations (due to intermittent failures during previous clean up)
tablesToCleanup.forEach(f -> {
try {
fs.delete(new Path(zindexFolderPath, f), true);
} catch (IOException ie) {
// NOTE: Exception is deliberately swallowed to not affect overall clustering operation,
// since failing Z-index table will be attempted to be cleaned up upon subsequent
// clustering iteration
LOG.warn(String.format("Failed to cleanup residual Z-index table: %s", f), ie);
}
});
} catch (IOException e) {
LOG.error("Failed to build new Z-index table", e);
throw new HoodieException("Failed to build new Z-index table", e);
}
}summary
Read:
- When the query type is optimized reading, both cow and mor can use zorder index optimization. When the query type is snapshot, only cow can
- scan files = all files - files that use zindex data but fail to hit
write in:
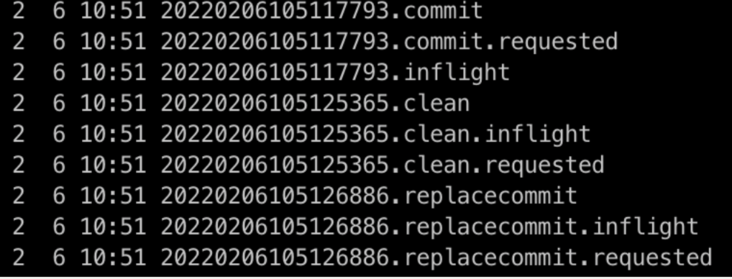
- It does not affect the writing of the main process of the task. The above figure 20220206105117793 is generated after the data is written and committed commit, other consumers can query normally
- The subsequent sorting and index generation can be executed synchronously after each write, or a scheduler task can be executed asynchronously in the background. This test inline cluster after each write, and finally 20220206105126886 replacecommit instant
relevant
iceberg's metadata counts the Min/Max of each file column, so it only needs to reorder the data in implementation, and there are relevant pr in the near future Spark: Spark3 ZOrder Rewrite Strategy . It is still an Action that needs to be triggered manually. The sorting strategy of Zorder will be added. A special article will be written to explain the specific details. It is mainly to be familiar with the differences between iceberg architecture and hudi architecture