Are you tired of GO and KEGG? Try consusupathdb. Its official website is: http://cpdb.molgen.mpg.de/
Interestingly, it compares 31 existing databases of biological functional gene sets and makes an arrangement. Is it an integrator? include:
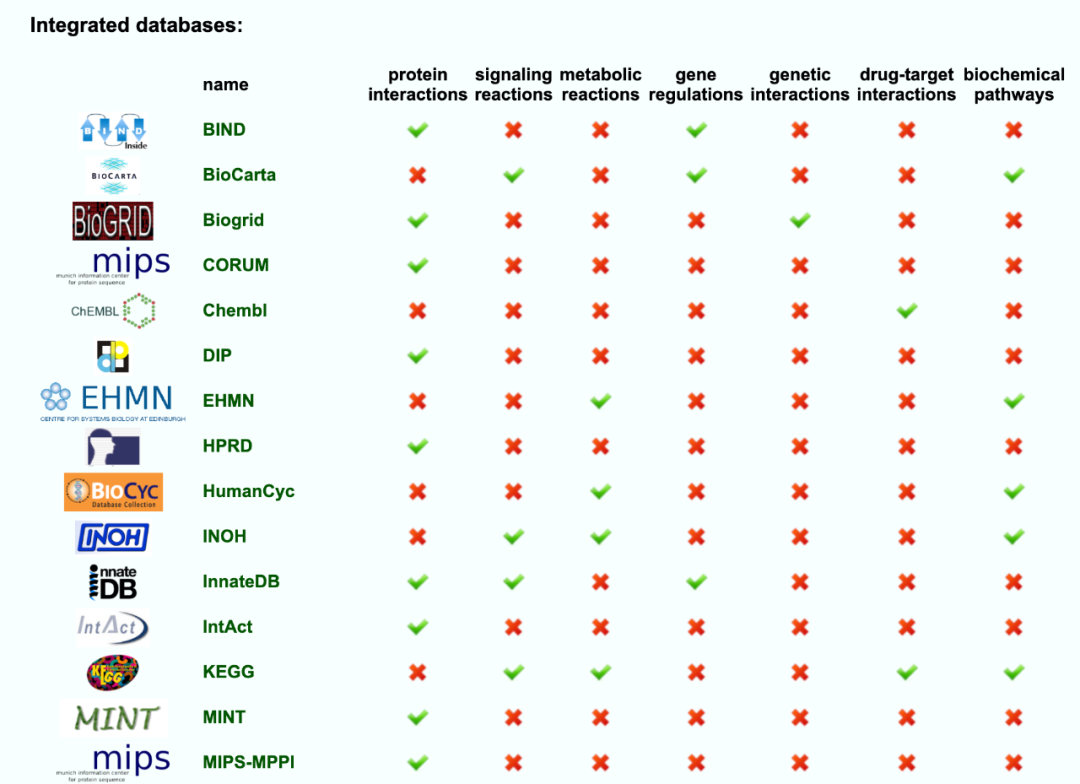
31 existing databases of biological functional gene sets
After a brief look, I'm familiar with KEGG database. The author said that this data set is the integrator of these other database resources, including:
- protein interactions
- signaling reactions
- metabolic reactions
- gene regulations
- genetic interactions
- drug-target interactions
- biochemical pathways
At present, there are more than 5000 gene sets. The details are as follows:
unique physical entities: | 200,499 |
|---|---|
unique interactions: | 859,848 |
gene regulations: | 18,912 |
protein interactions: | 616,304 |
genetic interactions: | 7,936 |
biochemical reactions: | 25,046 |
drug-target interactions: | 191,650 |
pathways: | 5,578 |
Each update has an article published, from 2009 to 2016:
Herwig, R. et al.. Analyzing and interpreting genome data at the network level with ConsensusPathDB. (2016) Nature Protocols 11, 1889-1907.Kamburov, A. et al. (2013) The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res.Kamburov, A. et al. (2011) ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res.Kamburov, A. et al. (2009) ConsensusPathDB--a database for integrating human interaction networks. Nucleic Acids Res.Pentchev, K. et al. (2010) Evidence mining and novelty assessment of protein-protein interactions with the ConsensusPathDB plugin for Cytoscape. Bioinformatics
At present, it has been updated to Release 35 (05.06.2021), and only the three species of human yeast mouse are supported temporarily.
Support downloading all more than 5000 gene sets
The gene set sorted out by the author has multiple IDS, including:
- Entrez
- Ensembl
- symbol (HGNC symbol)
- HGNC ID
- RefSeq
- Unigene
- Uniprot
I usually choose symbol (HGNC symbol), as shown below:
Butanoate metabolism - Homo sapiens (human) path:hsa00650 KEGG ACADS,ABAT,AACS,GAD1,GAD2,ACSM1,EHHADH,ACSM3,HADHA,ACSM5,ACSM4,ACSM6,ACAT1,ACAT2,HMGCLL1,HMGCS1,HMGCS2,HMGCL,ACSM2A,ACSM2B,BDH2,BDH1,HADH,L2HGDH,ECHS1,OXCT1,OXCT2,ALDH5A1 Steroid hormone biosynthesis - Homo sapiens (human) path:hsa00140 KEGG HSD17B12,HSD17B1,CYP11B2,HSD11B1L,UGT2A3,UGT2A2,UGT2A1,CYP17A1,UGT1A8,UGT1A9,UGT1A4,UGT1A5,UGT1A6,UGT1A7,UGT1A1,AKR1C2,UGT1A3,CYP7B1,CYP11A1,CYP1B1,UGT1A10,AKR1C4,CYP3A7-CYP3A51P,HSD17B8,UGT2B15,UGT2B17,UGT2B10,UGT2B11,CYP21A2,SRD5A3,SRD5A2,SRD5A1,AKR1C3,HSD11B2,HSD17B6,HSD17B7,UGT2B7,UGT2B4,AKR1C1,HSD3B2,DHRS11,LRTOMT,COMT,STS,SULT1E1,CYP3A7,CYP3A4,CYP3A5,HSD17B2,CYP1A1,CYP1A2,HSD17B3,CYP19A1,AKR1D1,CYP2E1,HSD3B1,SULT2B1,UGT2B28,CYP11B1,HSD11B1,CYP7A1 Cell cycle - Homo sapiens (human) path:hsa04110 KEGG BUB1B,PLK1,PKMYT1,GADD45A,GADD45B,ANAPC11,ANAPC10,ANAPC13,GADD45G,ESPL1,WEE2,WEE1,CUL1,CDKN2B,CDKN2C,CDKN2A,CDKN2D,MDM2,PRKDC,CCND1,CCND3,CCND2,GSK3B,SFN,DBF4,RBX1,CCNH,FZR1,CDKN1C,CDKN1B,CDKN1A,CDC16,CCNE2,CCNE1,TTK,STAG2,CHEK2,CHEK1,SMC1A,SMC1B,BUB1,BUB3,CDC25C,CDC25B,CDC25A,STAG1,ANAPC1,ZBTB17,ANAPC2,ANAPC5,ANAPC4,ANAPC7,TP53,CDC27,CDC26,CDC23,CDC14B,CDC14A,CDC20,TFDP2,TFDP1,HDAC1,HDAC2,SMAD4,ATM,SMAD2,SMAD3,MAD2L1,ATR,ABL1,CDC6,CDC7,SKP2,SKP1,SMC3,PTTG1,PTTG2,RBL1,RBL2,E2F5,EP300,MYC,CDK1,CDK2,CDK4,CDK6,CDK7,CDC45,MAD2L2,TGFB1,TGFB2,TGFB3,YWHAZ,CCNA2,CCNA1,MAD1L1,YWHAQ,RB1,YWHAH,YWHAB,YWHAG,YWHAE,PCNA,RAD21,ORC6,ORC4,ORC5,ORC2,ORC3,ORC1,MCM7,MCM6,MCM5,MCM4,MCM3,MCM2,CCNB3,CCNB2,CCNB1,E2F4,E2F3,E2F2,E2F1,CREBBP
As you can see, I randomly selected two gene sets, which were simply copied and pasted from KEGG database? So is the special value of this consumuspathdb to help us screen gene sets?
If you don't know R code, you also want to use this database
First of all, its own home page can support users to upload their own gene list. Official website: http://cpdb.molgen.mpg.de/
Secondly, it provides a Cytoscape plug-in, with the consumuspathdb plug-in for Cytoscape you can mine evidence (publications, detection methods, pathways, etc.) for interactions loaded in Cytoscape and highlight interactions that have not been detected previously Please read our paper on the plugin to learn how to use it.
If you really think my tutorial is helpful to your scientific research project and makes you enlightened, or your project uses a lot of my skills, please add a short thank you when publishing your achievements in the future, as shown below:
We thank Dr.Jianming Zeng(University of Macau), and all the members of his bioinformatics team, biotrainee, for generously sharing their experience and codes.
If I have traveled around the world in universities and research institutes (including Chinese mainland) in ten years, I will give priority to seeing you if you have such a friendship.