
Holding fireworks to make a living, poetic to seek love
catalogue
2.2 data center (Standardization)
2.4 calculate eigenvalues and eigenvectors of covariance matrix
2.5 determining the number of principal components
2.6 calculation of principal components
3.5 calculation of characteristic value
3.6 calculate the eigenvector corresponding to the eigenvalue
3.7 determine the number of principal components and calculate the principal component matrix
3.8 calculation of principal components (data after dimensionality reduction)
4.2 main category pcamain java
This article is written in the learning process. Please correct any mistakes.
1. Principle introduction
In many scenarios, multivariable data need to be observed, which increases the workload of data collection to a certain extent. More importantly, there may be correlation between multiple variables, which increases the complexity of problem analysis. If each index is analyzed separately, the analysis results are often isolated and can not make full use of the information in the data. Therefore, blindly reducing the index will lose a lot of useful information and lead to wrong conclusions.
Therefore, we need to find a reasonable method to reduce the indicators to be analyzed and minimize the loss of information contained in the original indicators, so as to achieve the purpose of comprehensive analysis of the collected data. Because there is a certain correlation between the variables, we can consider changing the closely related variables into as few new variables as possible to make these new variables irrelevant, so we can use fewer comprehensive indicators to represent all kinds of information in each variable.
Principal component analysis is one of the most commonly used unsupervised dimensionality reduction methods. It is a statistical analysis method that transforms multiple variables into a few principal components through dimensionality reduction technology. These principal components can reflect most of the information of the original variables. They are usually expressed as some linear combination of the original variables.
2. Detailed steps
2.1 data acquisition
Suppose there is a set of data, m pieces of data, and each data has n evaluation indicators, which constitutes the original data matrix of m*n, that is, x, and the data corresponding to each variable is recorded as x1, X2, X3 Xn.
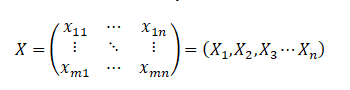
2.2 data center (Standardization)
Different evaluation indicators often have different dimensions and dimensional units, which will affect the results of data analysis. In order to eliminate the dimensional impact between indicators, data standardization is needed to solve the comparability between data indicators. After data standardization, all indicators of the original data are in the same order of magnitude, which is suitable for comprehensive comparative evaluation.
Here, we use the zero mean method (z-score) to process the data, and get the data subject to standard normal distribution with mean value of 0 and standard deviation of 1.
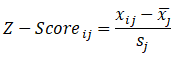
Among them,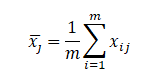 Represents the sample mean value of the j-th index,
Represents the sample mean value of the j-th index,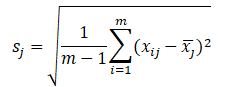
Represents the standard deviation of the j-th index, and the data matrix after centralization is still recorded as X.
2.3 finding covariance matrix
Calculate the covariance matrix of the centralized data and record it as R, then
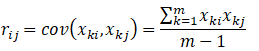
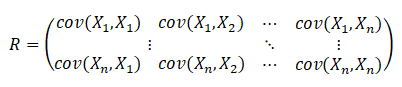
Or another way:
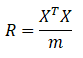
Calculation of eigenvalue and covariance of matrix 2.4
By solving the characteristic equation of the covariance matrix:
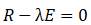
The eigenvalues are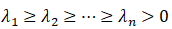
The corresponding eigenvectors are:
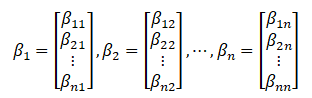
2.5 determining the number of principal components
Set a contribution rate threshold, that is, when the cumulative contribution rate of the characteristic values of the first p principal components is higher than this value, it can be considered that these p principal components can represent the original n variables, generally 0.8, 0.85, 0.9, 0.95, 0.99, etc.
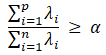
2.6 calculation of principal components
After the number of principal components is obtained, the eigenvectors corresponding to the first p eigenvalues can be used to calculate the principal components (dimension reduced data).
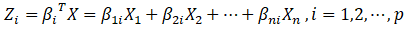
3. Case analysis
3.1 topic introduction
It is known that when judging the water quality of a certain water area, it can be judged by x1-x9 a total of 9 indexes. The index measurement of 11 rivers in A-K has brought great trouble in judging and determining the importance of each index due to too many indexes, Therefore, please use certain mathematical methods to reduce the evaluation indicators on the premise of trying not to lose the original data information.
| x1 | x2 | x3 | x4 | x5 | x6 | x7 | x8 | x9 | |
| A | 88 | 63 | 17 | 50 | 53 | 49 | 64 | 24 | 32 |
| B | 75 | 1 | 2 | 43 | 14 | 53 | 31 | 18 | 42 |
| C | 79 | 48 | 73 | 62 | 43 | 48 | 72 | 76 | 98 |
| D | 73 | 95 | 9 | 34 | 20 | 46 | 54 | 4 | 59 |
| E | 36 | 70 | 53 | 5 | 100 | 6 | 74 | 25 | 43 |
| F | 40 | 46 | 92 | 60 | 68 | 19 | 2 | 76 | 8 |
| G | 20 | 78 | 35 | 98 | 83 | 70 | 35 | 58 | 1 |
| H | 67 | 29 | 38 | 3 | 32 | 27 | 3 | 47 | 75 |
| I | 38 | 30 | 28 | 72 | 46 | 20 | 50 | 42 | 74 |
| J | 95 | 25 | 20 | 42 | 13 | 54 | 56 | 54 | 82 |
| K | 40 | 32 | 27 | 14 | 22 | 59 | 6 | 81 |
3.2 reading data
Using jxl package to read data from Excel and output it
//Read data
public double[][] read(String filepath) throws IOException, BiffException,WriteException {
//Create input stream
InputStream stream = new FileInputStream(filepath);
//Get Excel file object
Workbook rwb = Workbook.getWorkbook(stream);
//Gets the first default of the specified worksheet of the file
Sheet sheet = rwb.getSheet("Sheet1");
rows = sheet.getRows();
cols = sheet.getColumns();
double[][] orig = new double[rows][cols];
//Row as row
for(int i=0;i<sheet.getRows();i++) {
for(int j=0;j<sheet.getColumns();j++) {
String[] str = new String[sheet.getColumns()];
Cell cell = null;
cell = sheet.getCell(j,i);
str[j] = cell.getContents();
orig[i][j] = Double.valueOf(str[j]);
//original.set(i, j, orig[i][j]);
}
}
return orig;
}Output:
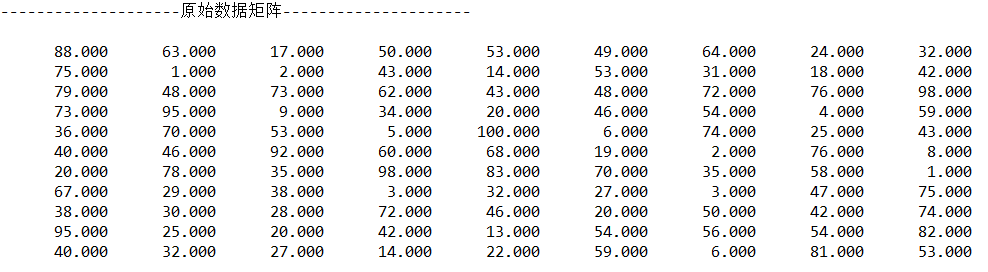
3.3 centralized data
The mean zeroing method is used to centralize the original data
/**
*
* Make the mean value of each sample 0
*
* @param primary
* Original two-dimensional array matrix
* @return averageArray Centralized matrix
*/
public double[][] changeAverageToZero(double[][] primary) {
int n = primary.length;
int m = primary[0].length;
double[] sum = new double[m];
double[] average = new double[m];
double[][] averageArray = new double[n][m];
for (int i = 0; i < m; i++) {
for (int j = 0; j < n; j++) {
sum[i] += primary[j][i];
}
average[i] = sum[i] / n;
}
for (int i = 0; i < m; i++) {
for (int j = 0; j < n; j++) {
averageArray[j][i] = primary[j][i] - average[i];
}
}
return averageArray;
}Output:
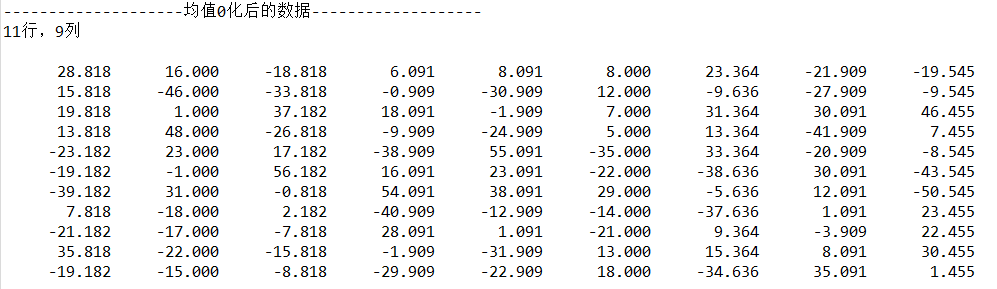
3.4 finding covariance matrix
/**
*
* Calculate covariance matrix
*
* @param matrix
* Centralized matrix
* @return result covariance matrix
*/
public double[][] getVarianceMatrix(double[][] matrix) {
int n = matrix.length;// Number of rows
int m = matrix[0].length;// Number of columns
double[][] result = new double[m][m];// covariance matrix
for (int i = 0; i < m; i++) {
for (int j = 0; j < m; j++) {
double temp = 0;
for (int k = 0; k < n; k++) {
temp += matrix[k][i] * matrix[k][j];
}
result[i][j] = temp / (n - 1);
}
}
/*Or use the following method to calculate: the transpose of X is multiplied by X and divided by the number of rows
Matrix X=new Matrix(matrix);
result = X.transpose().times(X).getArray();
for (int i = 0; i < m; i++) {
for (int j = 0; j < m; j++) {
result[i][j] = result[i][j] / n;
}
}
*/
return result;
}Output:
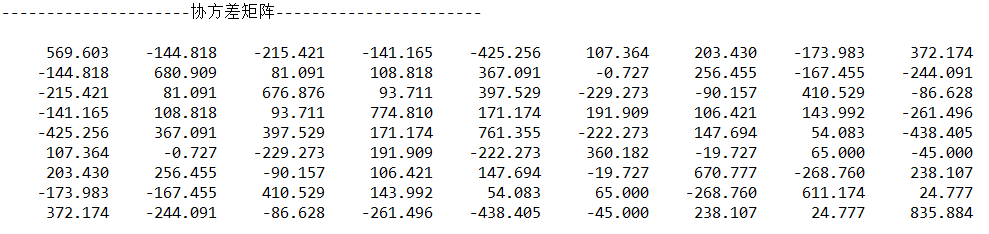
3.5 calculation of characteristic value
The data on the diagonal is the eigenvalue
/**
* Finding eigenvalue matrix
*
* @param matrix
* covariance matrix
* @return result Vector eigenvalue two-dimensional array matrix
*/
public double[][] getEigenvalueMatrix(double[][] matrix) {
Matrix A = new Matrix(matrix);
// A diagonal matrix composed of eigenvalues, which is obtained by eig()
// A.eig().getD().print(10, 6);
double[][] result = A.eig().getD().getArray();
return result;
}
Output:
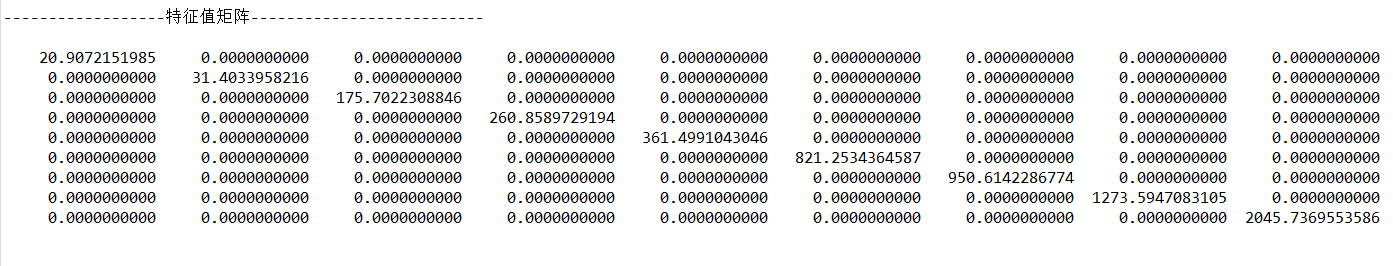
3.6 calculate the eigenvector corresponding to the eigenvalue
/**
* Normalized matrix (eigenvector matrix)
*
* @param matrix
* Eigenvalue matrix
* @return result Standardized two-dimensional array matrix
*/
public double[][] getEigenVectorMatrix(double[][] matrix) {
Matrix A = new Matrix(matrix);
// A.eig().getV().print(6, 2);
double[][] result = A.eig().getV().getArray();
return result;
}
Output:
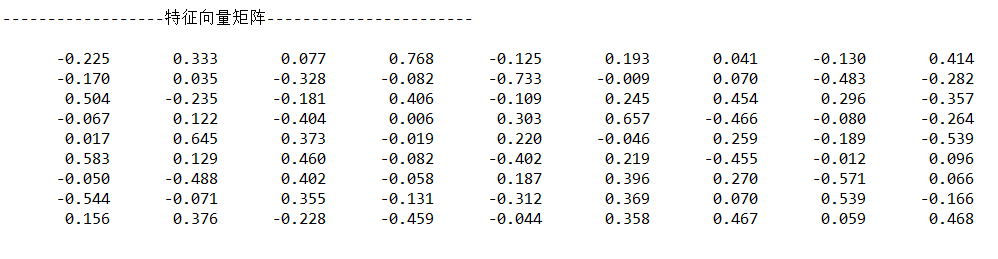
3.7 determine the number of principal components and calculate the principal component matrix
/**
* Looking for principal components
*
* @param prinmaryArray
* Original 2D array
* @param eigenvalue
* Two dimensional array of eigenvalues
* @param eigenVectors
* Two dimensional array of eigenvectors
* @return principalMatrix Principal component matrix
*/
public Matrix getPrincipalComponent(double[][] primaryArray,
double[][] eigenvalue, double[][] eigenVectors) {
Matrix A = new Matrix(eigenVectors);// Define an eigenvector
double[][] tEigenVectors = A.transpose().getArray();// Eigenvector transpose
Map<Integer, double[]> principalMap = new HashMap<Integer, double[]>();// key = principal component eigenvalue, value = eigenvector corresponding to the eigenvalue
TreeMap<Double, double[]> eigenMap = new TreeMap<Double, double[]>(
Collections.reverseOrder());// Key = eigenvalue, value = corresponding eigenvector; Initialize to flip sort, so that the map is arranged in descending order according to the key value
double total = 0;// Store sum of eigenvalues
int index = 0, n = eigenvalue.length;
double[] eigenvalueArray = new double[n];// Put the elements on the diagonal of the eigenvalue matrix into the array eigenvalueArray
for (int i = 0; i < n; i++) {
for (int j = 0; j < n; j++) {
if (i == j)
eigenvalueArray[index] = eigenvalue[i][j];
}
index++;
}
for (int i = 0; i < tEigenVectors.length; i++) {
double[] value = new double[tEigenVectors[0].length];
value = tEigenVectors[i];
eigenMap.put(eigenvalueArray[i], value);
}
// Sum of features
for (int i = 0; i < n; i++) {
total += eigenvalueArray[i];
}
// Select the first several principal components
double temp = 0;
int principalComponentNum = 0;// Principal component fraction
List<Double> plist = new ArrayList<Double>();// Principal component eigenvalue
for (double key : eigenMap.keySet()) {
if (temp / total <= threshold) {
temp += key;
plist.add(key);
principalComponentNum++;
}
}
System.out.println("\n" + "Current threshold: " + threshold);
System.out.println("Principal component score obtained: " + principalComponentNum + "\n");
System.out.println("The eigenvalue corresponding to the principal component (eigenvector) and its contribution rate are as follows:");
for (int i = 0; i<principalComponentNum; i++) {
System.out.println(plist.get(i)+"\t"+plist.get(i)*100/total+"%");
}
// Input data into the principal component map
for (int i = 0; i < plist.size(); i++) {
if (eigenMap.containsKey(plist.get(i))) {
principalMap.put(i, eigenMap.get(plist.get(i)));
}
}
// Save the values in the map into a two-dimensional array
double[][] principalArray = new double[principalMap.size()][];
Iterator<Entry<Integer, double[]>> it = principalMap.entrySet()
.iterator();
for (int i = 0; it.hasNext(); i++) {
principalArray[i] = it.next().getValue();
}
/*
double[][] principalArray1 = new double[principalArray.length][principalArray[0].length+1];
for(int i=0;i<principalArray1.length ;i++) {
for(int j=0;j<principalArray1.length ;j++) {
if(j==0) {
principalArray1[i][j] = plist.get(i);
}
else {
principalArray1[i][j] = principalArray[i][j-1];
}
}
}*/
Matrix principalMatrix = new Matrix(principalArray);
return principalMatrix;
}
Output:
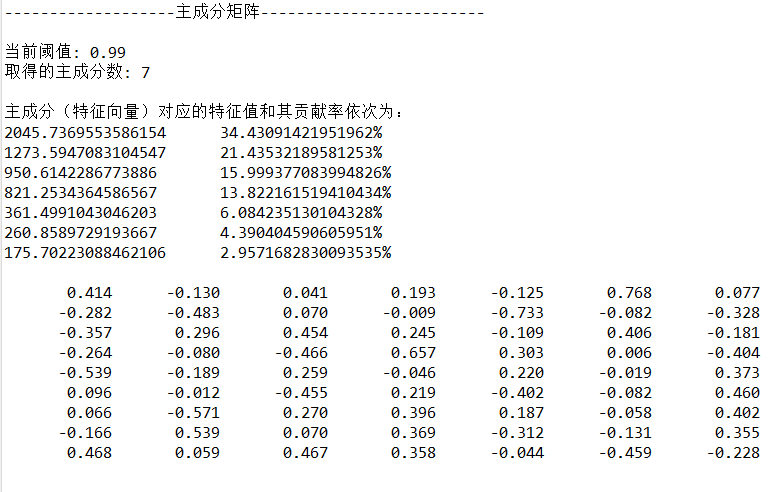
3.8 calculation of principal components (data after dimensionality reduction)
/**
* matrix multiplication
*
* @param primary
* Original 2D array
*
* @param matrix
* Principal component matrix
*
* @return result Result matrix
*/
public Matrix getResult(double[][] primary, Matrix matrix) {
Matrix primaryMatrix = new Matrix(primary);
Matrix result = primaryMatrix.times(matrix.transpose());
return result;
}Output:
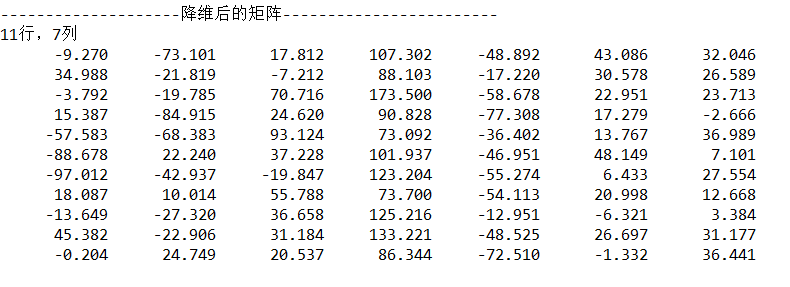
4. Complete code (Java)
The library for Excel data operation in the program is jxl and the matrix operation library is jama.
4.1 method PCA java
package PCA;
import java.io.FileInputStream;
import java.io.IOException;
import java.io.InputStream;
import java.util.ArrayList;
import java.util.Collections;
import java.util.HashMap;
import java.util.Iterator;
import java.util.List;
import java.util.Map;
import java.util.Map.Entry;
import java.util.TreeMap;
import Jama.Matrix;
import jxl.Cell;
import jxl.Sheet;
import jxl.Workbook;
import jxl.read.biff.BiffException;
import jxl.write.WriteException;
/*
* Algorithm steps:
* 1)Form the original data into an n-row m-column matrix X by column
* 2)Feature centralization. That is, the mean value of each dimension is subtracted from the data of each dimension, so that the mean value of each dimension is 0
* 3)Find the covariance matrix
* 4)The eigenvalues of the covariance matrix and the corresponding eigenvectors are obtained
* 5)The eigenvectors are arranged into a matrix from top to bottom according to the corresponding eigenvalue size, and the first k rows are taken to form the matrix p
* 6)Y=PX That is, the data after dimension reduction to k dimension
*/
public class pca {
private static final double threshold = 0.99;// Threshold characteristic value
int rows,cols;
//Output two-dimensional matrix
public void matrixoutput(double[][] x) {
for(int i=0;i<x.length;i++) {
for(int j=0;j<x[0].length;j++) {
System.out.print(x[i][j]+" ");
}
System.out.println();
}
}
//Read data
public double[][] read(String filepath) throws IOException, BiffException,WriteException {
//Create input stream
InputStream stream = new FileInputStream(filepath);
//Get Excel file object
Workbook rwb = Workbook.getWorkbook(stream);
//Gets the first default of the specified worksheet of the file
Sheet sheet = rwb.getSheet("Sheet1");
rows = sheet.getRows();
cols = sheet.getColumns();
double[][] orig = new double[rows][cols];
//Row as row
for(int i=0;i<sheet.getRows();i++) {
for(int j=0;j<sheet.getColumns();j++) {
String[] str = new String[sheet.getColumns()];
Cell cell = null;
cell = sheet.getCell(j,i);
str[j] = cell.getContents();
orig[i][j] = Double.valueOf(str[j]);
//original.set(i, j, orig[i][j]);
}
}
return orig;
}
/**
*
* Make the mean value of each sample 0
*
* @param primary
* Original two-dimensional array matrix
* @return averageArray Centralized matrix
*/
public double[][] changeAverageToZero(double[][] primary) {
int n = primary.length;
int m = primary[0].length;
double[] sum = new double[m];
double[] average = new double[m];
double[][] averageArray = new double[n][m];
for (int i = 0; i < m; i++) {
for (int j = 0; j < n; j++) {
sum[i] += primary[j][i];
}
average[i] = sum[i] / n;
}
for (int i = 0; i < m; i++) {
for (int j = 0; j < n; j++) {
averageArray[j][i] = primary[j][i] - average[i];
}
}
return averageArray;
}
/**
*
* Calculate covariance matrix
*
* @param matrix
* Centralized matrix
* @return result covariance matrix
*/
public double[][] getVarianceMatrix(double[][] matrix) {
int n = matrix.length;// Number of rows
int m = matrix[0].length;// Number of columns
double[][] result = new double[m][m];// covariance matrix
for (int i = 0; i < m; i++) {
for (int j = 0; j < m; j++) {
double temp = 0;
for (int k = 0; k < n; k++) {
temp += matrix[k][i] * matrix[k][j];
}
result[i][j] = temp / (n - 1);
}
}
/*Or use the following method to calculate: the transpose of X is multiplied by X and divided by the number of rows
Matrix X=new Matrix(matrix);
result = X.transpose().times(X).getArray();
for (int i = 0; i < m; i++) {
for (int j = 0; j < m; j++) {
result[i][j] = result[i][j] / n;
}
}
*/
return result;
}
/**
* Finding eigenvalue matrix
*
* @param matrix
* covariance matrix
* @return result Vector eigenvalue two-dimensional array matrix
*/
public double[][] getEigenvalueMatrix(double[][] matrix) {
Matrix A = new Matrix(matrix);
// A diagonal matrix composed of eigenvalues, which is obtained by eig()
// A.eig().getD().print(10, 6);
double[][] result = A.eig().getD().getArray();
return result;
}
/**
* Normalized matrix (eigenvector matrix)
*
* @param matrix
* Eigenvalue matrix
* @return result Standardized two-dimensional array matrix
*/
public double[][] getEigenVectorMatrix(double[][] matrix) {
Matrix A = new Matrix(matrix);
// A.eig().getV().print(6, 2);
double[][] result = A.eig().getV().getArray();
return result;
}
/**
* Looking for principal components
*
* @param prinmaryArray
* Original 2D array
* @param eigenvalue
* Two dimensional array of eigenvalues
* @param eigenVectors
* Two dimensional array of eigenvectors
* @return principalMatrix Principal component matrix
*/
public Matrix getPrincipalComponent(double[][] primaryArray,
double[][] eigenvalue, double[][] eigenVectors) {
Matrix A = new Matrix(eigenVectors);// Define an eigenvector
double[][] tEigenVectors = A.transpose().getArray();// Eigenvector transpose
Map<Integer, double[]> principalMap = new HashMap<Integer, double[]>();// key = principal component eigenvalue, value = eigenvector corresponding to the eigenvalue
TreeMap<Double, double[]> eigenMap = new TreeMap<Double, double[]>(
Collections.reverseOrder());// Key = eigenvalue, value = corresponding eigenvector; Initialize to flip sort, so that the map is arranged in descending order according to the key value
double total = 0;// Store sum of eigenvalues
int index = 0, n = eigenvalue.length;
double[] eigenvalueArray = new double[n];// Put the elements on the diagonal of the eigenvalue matrix into the array eigenvalueArray
for (int i = 0; i < n; i++) {
for (int j = 0; j < n; j++) {
if (i == j)
eigenvalueArray[index] = eigenvalue[i][j];
}
index++;
}
for (int i = 0; i < tEigenVectors.length; i++) {
double[] value = new double[tEigenVectors[0].length];
value = tEigenVectors[i];
eigenMap.put(eigenvalueArray[i], value);
}
// Sum of features
for (int i = 0; i < n; i++) {
total += eigenvalueArray[i];
}
// Select the first several principal components
double temp = 0;
int principalComponentNum = 0;// Principal component fraction
List<Double> plist = new ArrayList<Double>();// Principal component eigenvalue
for (double key : eigenMap.keySet()) {
if (temp / total <= threshold) {
temp += key;
plist.add(key);
principalComponentNum++;
}
}
System.out.println("\n" + "Current threshold: " + threshold);
System.out.println("Principal component score obtained: " + principalComponentNum + "\n");
System.out.println("The eigenvalue corresponding to the principal component (eigenvector) and its contribution rate are as follows:");
for (int i = 0; i<principalComponentNum; i++) {
System.out.println(plist.get(i)+"\t"+plist.get(i)*100/total+"%");
}
// Input data into the principal component map
for (int i = 0; i < plist.size(); i++) {
if (eigenMap.containsKey(plist.get(i))) {
principalMap.put(i, eigenMap.get(plist.get(i)));
}
}
// Save the values in the map into a two-dimensional array
double[][] principalArray = new double[principalMap.size()][];
Iterator<Entry<Integer, double[]>> it = principalMap.entrySet()
.iterator();
for (int i = 0; it.hasNext(); i++) {
principalArray[i] = it.next().getValue();
}
/*
double[][] principalArray1 = new double[principalArray.length][principalArray[0].length+1];
for(int i=0;i<principalArray1.length ;i++) {
for(int j=0;j<principalArray1.length ;j++) {
if(j==0) {
principalArray1[i][j] = plist.get(i);
}
else {
principalArray1[i][j] = principalArray[i][j-1];
}
}
}*/
Matrix principalMatrix = new Matrix(principalArray);
return principalMatrix;
}
/**
* matrix multiplication
*
* @param primary
* Original 2D array
*
* @param matrix
* Principal component matrix
*
* @return result Result matrix
*/
public Matrix getResult(double[][] primary, Matrix matrix) {
Matrix primaryMatrix = new Matrix(primary);
Matrix result = primaryMatrix.times(matrix.transpose());
return result;
}
}4.2 main category pcamain java
package PCA;
import Jama.Matrix;
import jxl.read.biff.BiffException;
import jxl.write.WriteException;
import java.io.IOException;
public class pcamain {
public static void main(String[] args) throws IOException, BiffException, WriteException {
// TODO Auto-generated catch block
//SelectData selectData = new SelectData();
pca pca = new pca();
//Get sample set
double[][] primaryArray = pca.read("pca.xls");
System.out.println("--------------------Raw data matrix---------------------");
//pca.matrixoutput(primaryArray);
Matrix A=new Matrix(primaryArray);
A.print(10, 3);
double[][] averageArray = pca.changeAverageToZero(primaryArray);
System.out.println();
System.out.println("--------------------Data after mean zeroing-------------------");
System.out.println(averageArray.length + "that 's ok," + averageArray[0].length + "column");
//pca.matrixoutput(averageArray);
Matrix B=new Matrix(averageArray);
B.print(10, 3);
System.out.println();
System.out.println("---------------------covariance matrix -----------------------");
double[][] varMatrix = pca.getVarianceMatrix(averageArray);
//pca.matrixoutput(varMatrix);
Matrix C=new Matrix(varMatrix);
C.print(10, 3);
System.out.println();
System.out.println("------------------Eigenvalue matrix--------------------------");
double[][] eigenvalueMatrix = pca.getEigenvalueMatrix(varMatrix);
//pca.matrixoutput(eigenvalueMatrix);
Matrix D=new Matrix(eigenvalueMatrix);
D.print(15, 10);
System.out.println();
System.out.println("------------------Eigenvector matrix-----------------------");
double[][] eigenVectorMatrix = pca.getEigenVectorMatrix(varMatrix);
//pca.matrixoutput(eigenVectorMatrix);
Matrix E=new Matrix(eigenVectorMatrix);
E.print(10, 3);
System.out.println();
System.out.println("-------------------Principal component matrix-------------------------");
Matrix principalMatrix = pca.getPrincipalComponent(primaryArray, eigenvalueMatrix, eigenVectorMatrix);
principalMatrix.transpose().print(10, 3);
System.out.println();
System.out.println("--------------------Reduced dimension matrix------------------------");
Matrix resultMatrix = pca.getResult(primaryArray, principalMatrix);
int c = resultMatrix.getColumnDimension(); //Number of columns
int r = resultMatrix.getRowDimension();//Number of rows
System.out.print(r + "that 's ok," + c + "column");
resultMatrix.print(10, 3);
}
}