preface
Part of the content is extracted from the training materials of shangsilicon Valley, dark horse and so on
1. Hadoop Archive
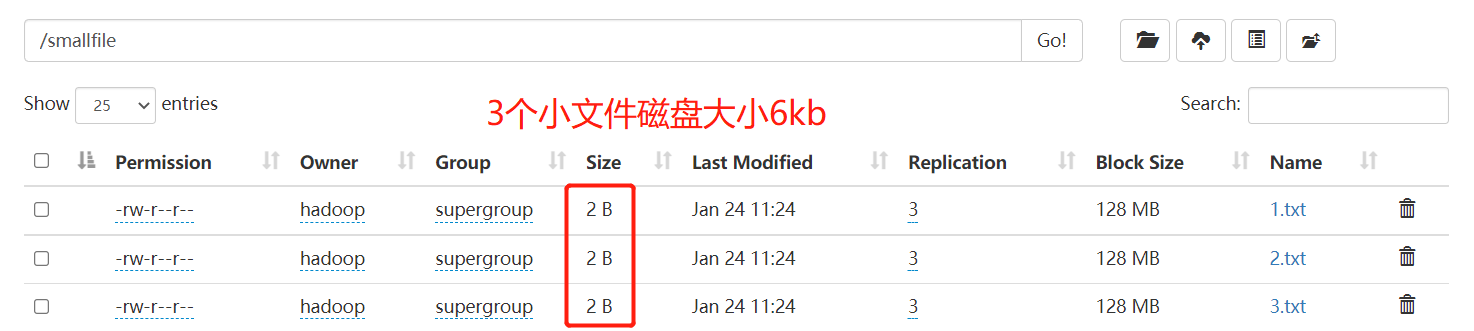
HDFS is not good at storing small files, because each file has at least one block, and the metadata of each block will occupy memory in the NameNode. If there are a large number of small files, they will eat up a large amount of memory in the NameNode. Simulate the small file scenario as follows:
[hadoop@hadoop1 input]$ hadoop fs -mkdir /smallfile [hadoop@hadoop1 input]$ echo 1 > 1.txt [hadoop@hadoop1 input]$ echo 2 > 2.txt [hadoop@hadoop1 input]$ echo 3 > 3.txt [hadoop@hadoop1 input]$ hadoop fs -put 1.txt 2.txt 3.txt /smallfile
Hadoop Archives can effectively deal with the above problems. It can archive multiple files into one file. After archiving into one file, it can also access each file transparently.
1.1 create Archive
Usage: hadoop archive -archiveName name -p <parent> <src>* <dest>
where - archiveName is the name of the archive to be created. For example, test Har, the extension of the archive name should be * har. - The p parameter specifies the relative path to the archive file (src).
for example: - p /foo/bar a/b/c e/f/g, where / foo/bar is the parent path of a/b/c and e/f/g, so the complete path is / foo/bar/a/b/c and / foo/bar/e/f/g.
for example, if you only want to archive all files in one directory / smallfile:
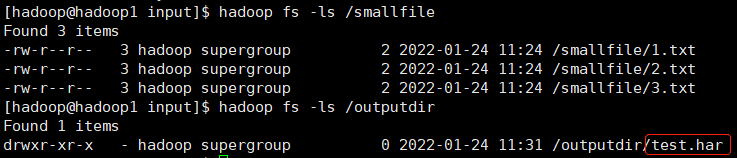
hadoop archive -archiveName test.har -p /smallfile /outputdir
in this way, a file named test. Will be created in the / outputdir directory Archive of har.
note: Archive archiving is completed through MapReduce program, and YARN cluster needs to be started.
1.2 view Archive
1.2.1 view the appearance after archiving
first, let's take a look at the created har file. Use the following command:
hadoop fs -ls /outputdir/test.har
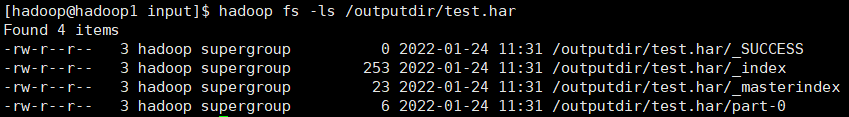
here you can see that the har file includes two index files, multiple part files (only one in this example) and a file that identifies whether it is successful or not. The part file is a collection of multiple original files. You can find the original file through the index file.
for example, the above three small files 1 txt 2. txt 3. Txt contents are 1, 2 and 3 respectively. After the archive operation, three small files are archived to test part-0 in Har is in a file.
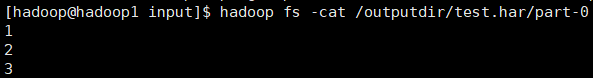
1.2.2 view what it looks like before archiving
when viewing the har file, if no access protocol is specified, the default is hdfs: / /, and what you can see is what it looks like after archiving.
in addition, Archive also provides its own har uri access protocol. If you use har uri to access, the index, identification and other files will be hidden, and only the original files before the file is created will be displayed:
the URI of Hadoop Archives is:
har://scheme-hostname:port/archivepath/fileinarchive
scheme hostname format is hdfs domain name: Port

hadoop fs -ls har://hdfs-node1:8020/outputdir/test.har/ hadoop fs -ls har:///outputdir/test.har hadoop fs -cat har:///outputdir/test.har/1.txt
1.3 extract Archive
decompress the archive in sequence (serial):
hadoop fs -cp har:///user/zoo/foo.har/dir1 hdfs:/user/zoo/newdir
hadoop fs -mkdir /smallfile1 hadoop fs -cp har:///outputdir/test.har/* /smallfile1 hadoop fs -ls /smallfile1
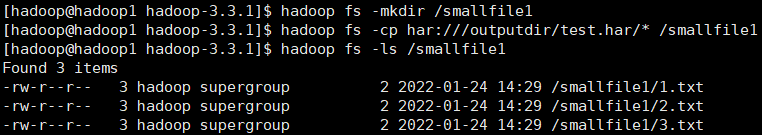
to decompress the archive in parallel, use DistCp. Corresponding to large archive files can improve efficiency:
hadoop distcp har:///user/zoo/foo.har/dir1 hdfs:/user/zoo/newdir
hadoop distcp har:///outputdir/test.har/* /smallfile2
1.4 precautions for using Archive
- Hadoop archives is a special file format. A Hadoop archive corresponds to a file system directory. The extension of Hadoop archive is * har;
- Creating archives is essentially running a Map/Reduce task, so you should run the command to create archives on the Hadoop cluster;
- Creating an archive file consumes as much hard disk space as the original file;
- Archive files do not support compression, although archive files appear to have been compressed;
- Once the archive file is created, it cannot be changed. If you want to modify it, you need to create a new archive file. In fact, the archived documents will not be modified because they are archived regularly, such as weekly or daily;
- When creating an archive, the source file will not be changed or deleted;
2. Sequence File
2.1 introduction to sequence file
Sequence File is a binary file support provided by Hadoop API. This binary file directly serializes the < key, value > key value pairs into the file.

2.2 advantages and disadvantages of sequence file
- advantage
- Secondary format storage, more compact than text files.
- Supports different levels of compression (based on Record or Block compression).
- Files can be split and processed in parallel, which is suitable for MapReduce.
- shortcoming
- Binary format files are inconvenient to view.
- Specific to hadoop, only the Java API can be used to interact with it. Multilingual support is not yet available.
2.3 Sequence File format
Hadoop Sequence File is a binary key / value pair. According to the compression type, there are three different Sequence File formats: uncompressed format, record compressed format and block compressed format.
Sequence File consists of one header and one or more record s. The above three formats all use the same header structure, as shown below:
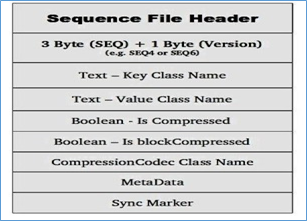
the first three bytes are SEQ, indicating that the file is a sequence file, followed by a byte indicating the actual version number (for example, SEQ 4 or SEQ 6). Other in the Header also include key, value, class name, compression details, metadata and Sync Marker. Sync Marker synchronization mark, which is used to read data at any position.
2.3.1 uncompressed format
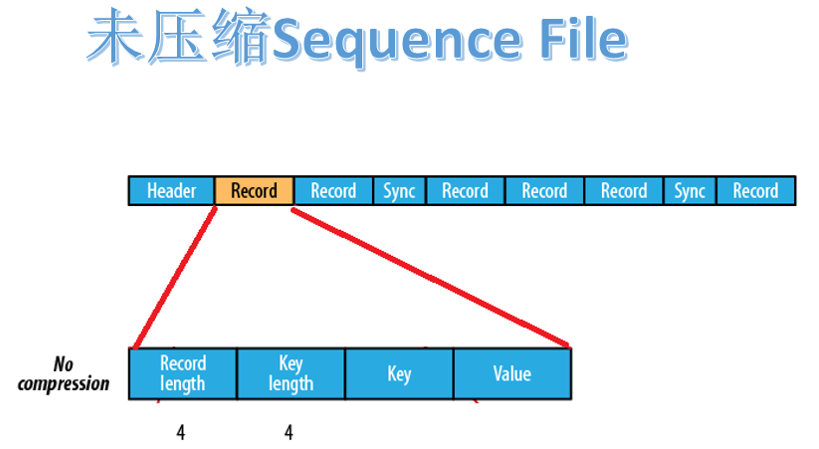
uncompressed Sequence File consists of header, record and sync. Record includes four parts: record length, key length, key and value.
there is a synchronization flag every few record s (about 100 bytes).
2.3.2 record based compression format
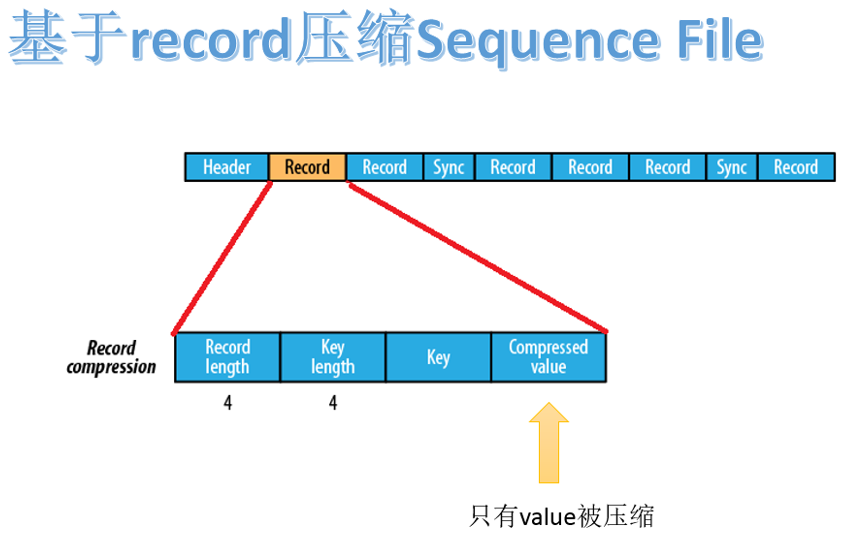
the Sequence File file compressed based on record is composed of header, record and sync. Record contains four parts: record length, key length, key and compressed value.
there is a synchronization flag every few record s (about 100 bytes).
2.3.3 block based compression format
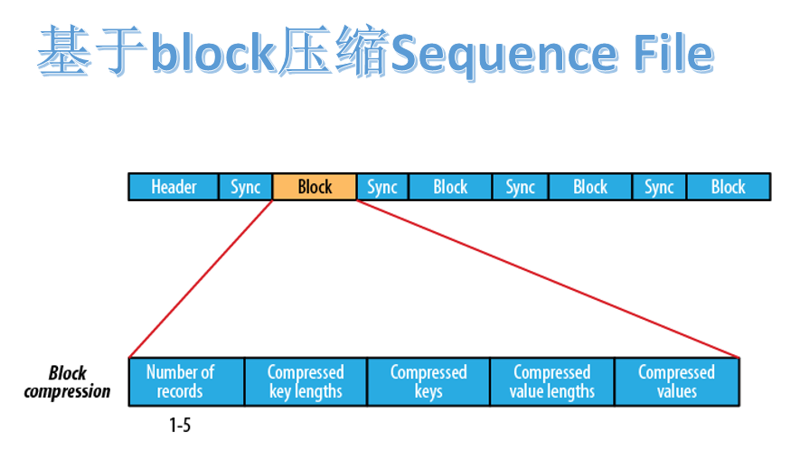
the Sequence File file based on block compression is composed of header, block and sync.
block refers to record block, which can be understood as a block composed of multiple record records. Note that this block is different from the block (128M) in HDFS.
block includes: number of record s, compressed key length, compressed keys, compressed value length and compressed values. Every other block has a synchronization flag.
block compression provides better compression ratio than record compression. When using Sequence File, block compression is usually preferred.
2.4 Sequence File reading and writing
2.4.1 construction of development environment
<dependencies> <dependency> <groupId>org.apache.hadoop</groupId> <artifactId>hadoop-common</artifactId> <version>3.3.1</version> </dependency> <dependency> <groupId>org.apache.hadoop</groupId> <artifactId>hadoop-hdfs</artifactId> <version>3.3.1</version> </dependency> <dependency> <groupId>org.apache.hadoop</groupId> <artifactId>hadoop-client</artifactId> <version>3.3.1</version> </dependency> </dependencies>
2.4.2 SequenceFileWrite
import java.net.URI;
import org.apache.hadoop.conf.Configuration;
import org.apache.hadoop.fs.FileSystem;
import org.apache.hadoop.fs.Path;
import org.apache.hadoop.io.IOUtils;
import org.apache.hadoop.io.IntWritable;
import org.apache.hadoop.io.SequenceFile;
import org.apache.hadoop.io.Text;
import org.apache.hadoop.io.compress.CompressionCodec;
import org.apache.hadoop.io.compress.GzipCodec;
public class SequenceFileWrite {
private static final String[] DATA = {
"One, two, buckle my shoe",
"Three, four, shut the door",
"Five, six, pick up sticks",
"Seven, eight, lay them straight",
"Nine, ten, a big fat hen"
};
public static void main(String[] args) throws Exception {
//Set the running identity of the client to operate as root to access HDFS
System.setProperty("HADOOP_USER_NAME","hadoop");
//Configuration is used to specify related parameter properties
Configuration conf = new Configuration();
//sequence file key,value
IntWritable key = new IntWritable();
Text value = new Text();
//Construct Writer parameter properties
SequenceFile.Writer writer = null;
CompressionCodec Codec = new GzipCodec();
SequenceFile.Writer.Option optPath = SequenceFile.Writer.file(new Path("hdfs://192.168.68.101:8020/seq.out"));
SequenceFile.Writer.Option optKey = SequenceFile.Writer.keyClass(key.getClass());
SequenceFile.Writer.Option optVal = SequenceFile.Writer.valueClass(value.getClass());
SequenceFile.Writer.Option optCom = SequenceFile.Writer.compression(SequenceFile.CompressionType.RECORD,Codec);
try {
writer = SequenceFile.createWriter( conf, optPath, optKey, optVal, optCom);
for (int i = 0; i < 100; i++) {
key.set(100 - i);
value.set(DATA[i % DATA.length]);
System.out.printf("[%s]\t%s\t%s\n", writer.getLength(), key, value);
writer.append(key, value);
}
} finally {
IOUtils.closeStream(writer);
}
}
}
operation results:

the final output documents are as follows:

2.4.3 SequenceFileRead
import org.apache.hadoop.conf.Configuration;
import org.apache.hadoop.fs.Path;
import org.apache.hadoop.io.IOUtils;
import org.apache.hadoop.io.SequenceFile;
import org.apache.hadoop.io.Writable;
import org.apache.hadoop.util.ReflectionUtils;
import java.io.IOException;
public class SequenceFileRead {
public static void main(String[] args) throws IOException {
//Set the client running identity to operate as root to access HDFS
System.setProperty("HADOOP_USER_NAME","hadoop");
//Configuration is used to specify related parameter properties
Configuration conf = new Configuration();
SequenceFile.Reader.Option option1 = SequenceFile.Reader.file(new Path("hdfs://192.168.68.101:8020/seq.out"));
SequenceFile.Reader.Option option2 = SequenceFile.Reader.length(174);//This parameter represents the read length
SequenceFile.Reader reader = null;
try {
reader = new SequenceFile.Reader(conf,option1,option2);
Writable key = (Writable) ReflectionUtils.newInstance(
reader.getKeyClass(), conf);
Writable value = (Writable) ReflectionUtils.newInstance(
reader.getValueClass(), conf);
long position = reader.getPosition();
while (reader.next(key, value)) {
String syncSeen = reader.syncSeen() ? "*" : "";//Is the Sync Mark sync flag returned
System.out.printf("[%s%s]\t%s\t%s\n", position, syncSeen, key, value);
position = reader.getPosition(); // beginning of next record
}
} finally {
IOUtils.closeStream(reader);
}
}
}
operation results:
2.5 case: merging small files using Sequence File
2.5.1 theoretical basis
you can use Sequence File to merge small files, that is, serialize the file name as key and the file content as value into a large file. For example, suppose there are 10000 100KB files, we can write a program to put them into a single Sequence File, as shown below, where we can use filename as the key and content as the value.
2.5.2 specific values
import java.io.File;
import java.io.FileInputStream;
import java.nio.charset.Charset;
import java.util.ArrayList;
import java.util.List;
import org.apache.commons.codec.digest.DigestUtils;
import org.apache.hadoop.conf.Configuration;
import org.apache.hadoop.fs.FileUtil;
import org.apache.hadoop.fs.Path;
import org.apache.hadoop.io.BytesWritable;
import org.apache.hadoop.io.SequenceFile;
import org.apache.hadoop.io.SequenceFile.Reader;
import org.apache.hadoop.io.SequenceFile.Writer;
import org.apache.hadoop.io.Text;
import org.slf4j.Logger;
import org.slf4j.LoggerFactory;
public class MergeSmallFilesToSequenceFile {
private Configuration configuration = new Configuration();
private List<String> smallFilePaths = new ArrayList<String>();
//Define the path that the method uses to add small files
public void addInputPath(String inputPath) throws Exception{
File file = new File(inputPath);
//If the given path is a folder, traverse the folder and put all the files in the subfolder into smallFilePaths
//If the given path is a file, put the path of the file into smallFilePaths
if(file.isDirectory()){
File[] files = FileUtil.listFiles(file);
for(File sFile:files){
smallFilePaths.add(sFile.getPath());
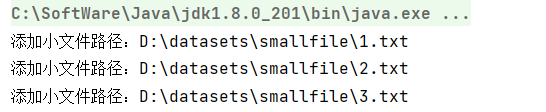
System.out.println("Add small file path:" + sFile.getPath());
}
}else{
smallFilePaths.add(file.getPath());
System.out.println("Add small file path:" + file.getPath());
}
}
//Traverse and read the small file of smallFilePaths, and then put it into the merged sequencefile container
public void mergeFile() throws Exception{
Writer.Option bigFile = Writer.file(new Path("D:\\datasets\\bigfile"));
Writer.Option keyClass = Writer.keyClass(Text.class);
Writer.Option valueClass = Writer.valueClass(BytesWritable.class);
//Construct writer
Writer writer = SequenceFile.createWriter(configuration, bigFile, keyClass, valueClass);
//Traverse and read small files and write sequencefile one by one
Text key = new Text();
for(String path:smallFilePaths){
File file = new File(path);
long fileSize = file.length();//Gets the number of bytes and size of the file
byte[] fileContent = new byte[(int)fileSize];
FileInputStream inputStream = new FileInputStream(file);
inputStream.read(fileContent, 0, (int)fileSize);//Load the binary stream of the file into the fileContent byte array
String md5Str = DigestUtils.md5Hex(fileContent);
System.out.println("merge Small file:"+path+",md5:"+md5Str);
key.set(path);
//Put the file path as the key and the file content as the value into the sequencefile
writer.append(key, new BytesWritable(fileContent));
}
writer.hflush();
writer.close();
}
//Read small files from large files
public void readMergedFile() throws Exception{
Reader.Option file = Reader.file(new Path("D:\\bigfile.seq"));
Reader reader = new Reader(configuration, file);
Text key = new Text();
BytesWritable value = new BytesWritable();
while(reader.next(key, value)){
byte[] bytes = value.copyBytes();
String md5 = DigestUtils.md5Hex(bytes);
String content = new String(bytes, Charset.forName("GBK"));
System.out.println("Read file:"+key+",md5:"+md5+",content:"+content);
}
}
public static void main(String[] args) throws Exception {
MergeSmallFilesToSequenceFile msf = new MergeSmallFilesToSequenceFile();
//Merge small files
msf.addInputPath("D:\\datasets\\smallfile");
msf.mergeFile();
//Read large files
// msf.readMergedFile();
}
}