With the continuous decline of the price of ngs, the transcriptome sequencing project has long entered the era of large samples. Of course, there was such a trend as early as the chip price was close to the people. At present, the price of single-cell transcriptome is also following this old path.
Then, for the transcriptome of large sample cohort, there is often no known reasonable grouping. At this time, it will be artificially grouped to see the heterogeneity of the cohort, such as grouping according to the level of immunity.
There are many ways to implement this grouping according to the immune level. Let's briefly demonstrate the hierarchical clustering of PCA and heat map and the scoring grouping such as gsea or gsva to see if there is any difference.
First look at the hierarchical clustering of heat maps
Need to load step1 output The expression matrix in rdata file, oh, if you don't know step1 output Rdata if you get it, look at the code at the end of the text.
Firstly, the expression matrix of the target gene set is selected for heat map and hierarchical clustering, and then simple violence grouping is carried out;
load(file = 'step1-output.Rdata')
cg=c('CD3D','CD3G CD247','IFNG','IL2RG','IRF1','IRF4','LCK','OAS2,STAT1')
cg
cg=cg[cg %in% rownames(dat)]
library(pheatmap)
p=pheatmap(dat[cg,])
hc=cutree(p$tree_col,5)
ac=data.frame(hc=as.character(hc))
rownames(ac)=colnames(dat)
pheatmap(dat[cg,],annotation_col = ac)
The results are as follows:
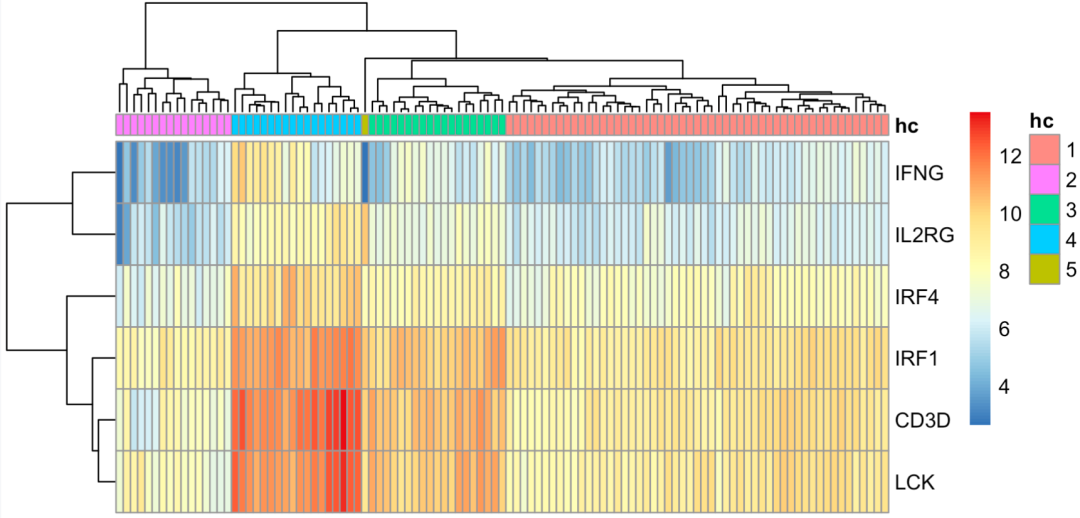
Hierarchical clustering
It can be seen that 1 and 2 are on the left and right sides of the heat map, while 3, 4 and 5 are in the middle. There is actually one sample in five groups. Therefore, we need to adjust the violence group to a reasonable immune gene group. The code is as follows:
group_list=ifelse(hc <3 ,'low','high') table(group_list) ac=data.frame(group_list) rownames(ac)=colnames(dat) pheatmap(dat[cg,],annotation_col = ac)
At this time, it can be seen that the samples are clearly divided into two groups:
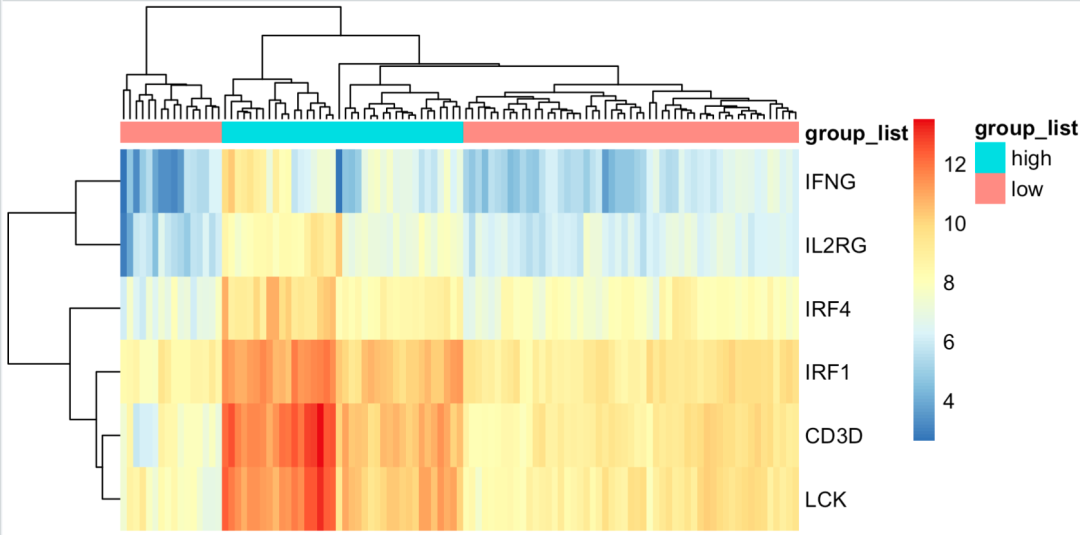
Hierarchical clustering and reasonable grouping
However, the number of such groups is not equal!
> table(group_list) group_list high low 38 69
It is worth mentioning that such high-low grouping of immune genes is a high-low concept within a data set, and cannot be merged across the data set.
We can also look at the expression matrix of this immune gene set by PCA. In fact, it is difficult to distinguish the global expression matrix after high and low grouping, but there is no problem in the small expression matrix of this immune gene set, as shown below:
library("FactoMineR") #These two packages need to be loaded to draw the principal component analysis diagram
library("factoextra")
dat.pca <- PCA(as.data.frame(t(dat[cg,])) )
fviz_pca_ind(dat.pca,
geom.ind = "point", # show points only (nbut not "text")
col.ind = group_list, # color by groups
addEllipses = T,
legend.title = "Groups"
)
Clearly visible boundaries of the two groups:
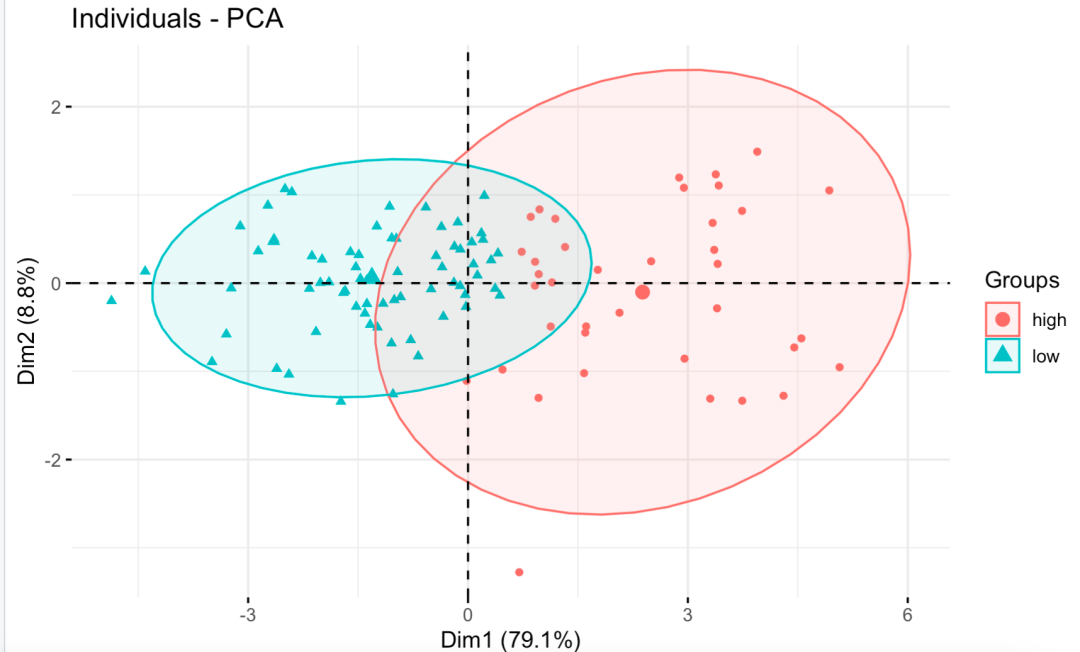
Clear difference boundary between the two groups of high and low immunity
About step1 output Rdata this file
The above code loads step1 - output Rdata is a file. Here is how to make this file. The code is as follows:
rm(list = ls()) ## Magic operation, one key emptying~
options(stringsAsFactors = F)
library(AnnoProbe)
library(GEOquery)
gset <- geoChina("GSE58812")
gset
gset[[1]]
a=gset[[1]] #
dat=exprs(a) #Now we need to know that expa is an object through the description of this function
dim(dat)#Look at the dimension of dat matrix
# GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array
dat[1:4,1:4] #Look at dat the 1 to 4 rows and 1 to 4 columns of this matrix. The row before the comma and the column after the comma
boxplot(dat[,1:4],las=2)
dat=log2(dat)
boxplot(dat[,1:4],las=2)
library(limma)
dat=normalizeBetweenArrays(dat)
boxplot(dat[,1:4],las=2)
pd=pData(a)
#By looking at the instruction manual, you can know to take the clinical information in object a and use pData
## Select some clinical phenotypes of interest.
library(stringr)
table(pd$`meta:ch1`)
group_list= ifelse(pd$`meta:ch1` == '0' ,'stable','meta')
table(group_list)
dat[1:4,1:4]
dim(dat)
a
ids=idmap('GPL570','soft')
head(ids)
ids=ids[ids$symbol != '',]
dat=dat[rownames(dat) %in% ids$ID,]
ids=ids[match(rownames(dat),ids$ID),]
head(ids)
colnames(ids)=c('probe_id','symbol')
ids$probe_id=as.character(ids$probe_id)
rownames(dat)=ids$probe_id
dat[1:4,1:4]
ids=ids[ids$probe_id %in% rownames(dat),]
dat[1:4,1:4]
dat=dat[ids$probe_id,]
ids$median=apply(dat,1,median) #ids creates a new column called media. At the same time, it operates the dat matrix by row, takes the median of each row, and gives the result to each row of the column media
ids=ids[order(ids$symbol,ids$median,decreasing = T),]#Sort ids$symbol according to the order of ids$median from large to small, and assign the corresponding row as a new ids
ids=ids[!duplicated(ids$symbol),]#Take out the duplicate item from the symbol column, '!' If it is no, that is, take out the non duplicate items, remove the duplicate gene, and retain the maximum expression result of each gene s
dat=dat[ids$probe_id,] #Remove the probe from the new ids_ In the column ID, the dat is formed into a new dat according to each row in the extracted column
rownames(dat)=ids$symbol#Give each row in the symbol column of ids to dat as the row name of dat
dat[1:4,1:4] #Keep the information of the first occurrence of each gene ID
dat['ACTB',]
dat['GAPDH',]
save(dat,group_list,
file = 'step1-output.Rdata')
If you really think my tutorial is helpful to your scientific research project and makes you enlightened, or your project uses a lot of my skills, please add a short thank you when publishing your achievements in the future, as shown below:
We thank Dr.Jianming Zeng(University of Macau), and all the members of his bioinformatics team, biotrainee, for generously sharing their experience and codes.
If I have traveled around the world in universities and research institutes (including Chinese mainland) in ten years, I will give priority to seeing you if you have such a friendship.