github address: DataScicence
Integrated learning 5-Xgboost principle and parameter tuning
Integrated learning 4-forward step-by-step algorithm and GBDT principle and case
Principles and cases of integrated learning 3-Boosting
Principle and case analysis of integrated learning 2-bagging
Principle and case analysis of integrated learning 1- voting method
Blending
principle
Steps:
- Divide data into trains_ data,Validate_data,Test_data three parts
- First layer model:
- Using multiple base models in Train_data to get multiple models M k M^k Mk
- Validate separately_ data,Test_ Data input model to get the prediction label A k , B k A^k,B^k Ak,Bk
- Layer 2 model:
- Use prediction labels on validation sets A k A^k Ak is used as the input and the real label is used as the output. The model is trained on the meta model N N N
- take B k B^k Bk is used as the input and N models are input to obtain the final prediction label on the test set T T T
- Evaluation model effect
Examples
import numpy as np import pandas as pd import matplotlib.pyplot as plt import seaborn as sns
Import data
from sklearn.datasets import load_iris
data = load_iris()
X = data.data[:,1:3] y = data.target print(X.shape,y.shape)
(150, 2) (150,)
Data division
from sklearn.model_selection import train_test_split X_train_,X_test,y_train_,y_test = train_test_split(X,y,test_size = 0.2) X_train,X_val,y_train,y_val = train_test_split(X_train_,y_train_)
print(X_train.shape,X_val.shape,X_test.shape)
(90, 2) (30, 2) (30, 2)
First layer classifier
from sklearn.svm import SVC from sklearn.tree import DecisionTreeClassifier from sklearn.neighbors import KNeighborsClassifier # Model M^k clfs = [SVC(probability = True),DecisionTreeClassifier(),KNeighborsClassifier()]
val_features = np.zeros((X_val.shape[0],len(clfs))) # A^k
test_features = np.zeros((X_test.shape[0],len(clfs))) #B^k
for i,clf in enumerate(clfs):
clf.fit(X_train,y_train)
val_feature = clf.predict_proba(X_val)[:, 1]
test_feature = clf.predict_proba(X_test)[:,1]
val_features[:,i] = val_feature
test_features[:,i] = test_feature
Second layer classifier
from sklearn.linear_model import LogisticRegression lr = LogisticRegression() #Model N lr.fit(val_features,y_val) # Output predicted results from sklearn.model_selection import cross_val_score cross_val_score(lr,test_features,y_test,cv=5)
array([0.83333333, 0.66666667, 0.66666667, 0.83333333, 0.66666667])
Draw decision boundary
The Blending boundary here is not accurate, because the input of lr model is the prediction result of the first layer model, not the training data
from mlxtend.plotting import plot_decision_regions
import matplotlib.gridspec as gridspec
import itertools
clfs.append(lr)
gs = gridspec.GridSpec(2, 2)
fig = plt.figure(figsize=(10,8))
for clf, lab, grd in zip(clfs,
['SVC',
'DT',
'KNN',
'Blending'],
itertools.product([0, 1], repeat=2)):
clf.fit(X, y)
ax = plt.subplot(gs[grd[0], grd[1]])
fig = plot_decision_regions(X=X, y=y, clf=clf)
plt.title(lab)
plt.show()

stacking
principle
In Blending, our training model only uses part of the data, and the data is not fully utilized. Therefore, we can use cross validation to use all the data
Steps:
- Segment data into training set Train_data, test set_ data
- First layer model
- K-fold cross validation is used on M models m respectively_ Data, and predict on K verification sets to get the prediction label A k ( take k individual junction fruit heap Fold , And Discipline Practice number according to of long degree mutually with ) A ^ k (stack K results with the same length as the training data) AK (stack k results with the same length as the training data), predict on the test set and obtain the prediction label B k ( take K individual junction fruit plus power flat all , And measure try collection long degree mutually with ) B ^ k (weighted average of K results, the same length as the test set) BK (weighted average of K results, the same length as the test set)
- The results of m models are stacked vertically A m × k , B m × k A^{m\times k},B^{m\times k} Am×k,Bm×k
- Second layer model
- use A m × k A^{m\times k} Am × k is used as input and trained on Meta model N to obtain model N
- take B m × k B^{m\times k} Bm × k input model to get the final prediction result
- Evaluation model effect
Examples
from sklearn.model_selection import cross_val_score from sklearn.linear_model import LogisticRegression from sklearn.neighbors import KNeighborsClassifier from sklearn.naive_bayes import GaussianNB from sklearn.ensemble import RandomForestClassifier from mlxtend.classifier import StackingCVClassifier
model training
clf1 = KNeighborsClassifier(n_neighbors=1)
clf2 = RandomForestClassifier()
clf3 = GaussianNB()
lr = LogisticRegression()
clf_stacking = StackingCVClassifier(classifiers=[clf1,clf2,clf3],
meta_classifier=lr,
cv=5)
for clf, label in zip([clf1, clf2, clf3, clf_stacking], ['KNN', 'Random Forest', 'Naive Bayes','StackingClassifier']):
scores = cross_val_score(clf, X, y, cv=3, scoring='accuracy')
print("Accuracy: %0.2f (+/- %0.2f) [%s]" % (scores.mean(), scores.std(), label))
Accuracy: 0.91 (+/- 0.01) [KNN] Accuracy: 0.95 (+/- 0.01) [Random Forest] Accuracy: 0.91 (+/- 0.02) [Naive Bayes] Accuracy: 0.96 (+/- 0.02) [StackingClassifier]
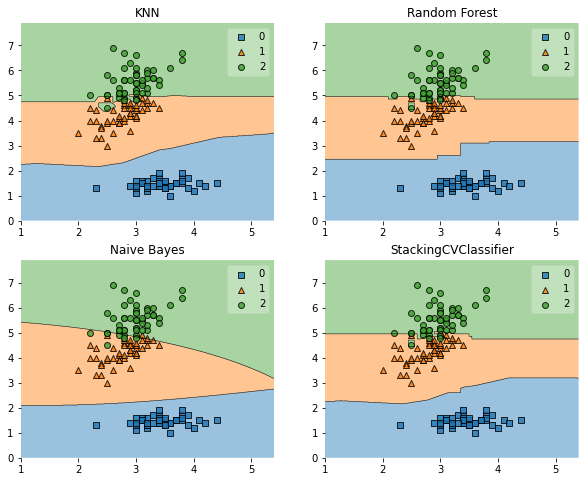
Decision boundary
from mlxtend.plotting import plot_decision_regions
from matplotlib import gridspec
import itertools
gs = gridspec.GridSpec(2,2)
fig = plt.figure(figsize=(10,8))
for clf,lab,grd in zip([clf1,clf2,clf3,clf_stacking],
['KNN',
'Random Forest',
'Naive Bayes',
'StackingCVClassifier'],
itertools.product([0,1],repeat=2)
):
clf.fit(X,y)
ax = plt.subplot(gs[grd[0],grd[1]])
fig = plot_decision_regions(X=X,y=y,clf=clf)
plt.title(lab)
plt.show()
Grid parameter adjustment of stacking model
from sklearn.model_selection import GridSearchCV
params = {'kneighborsclassifier__n_neighbors': [1, 5],
'randomforestclassifier__n_estimators': [10, 50],
'meta_classifier__C': [0.1, 10.0]}
grid = GridSearchCV(estimator=clf_stacking,
param_grid=params,
cv=5,
refit=True)
grid.fit(X, y)
print('Best parameters: %s' % grid.best_params_)
print('Accuracy: %.2f' % grid.best_score_)
Best parameters: {'kneighborsclassifier__n_neighbors': 5, 'meta_classifier__C': 0.1, 'randomforestclassifier__n_estimators': 50}
Accuracy: 0.95
clf_stacking.score(X,y)
0.9866666666666667
The base model uses different feature subsets
from sklearn.pipeline import make_pipeline
from mlxtend.feature_selection import ColumnSelector
iris = load_iris()
X = iris.data
y = iris.target
pipe1 = make_pipeline(ColumnSelector(cols=(0, 2)), # Select column 0,2
LogisticRegression())
pipe2 = make_pipeline(ColumnSelector(cols=(1, 2, 3)), # Select columns 1, 2 and 3
LogisticRegression())
sclf = StackingCVClassifier(classifiers=[pipe1, pipe2],
meta_classifier=LogisticRegression(),
random_state=42)
sclf.fit(X, y)
StackingCVClassifier(classifiers=[Pipeline(steps=[('columnselector',
ColumnSelector(cols=(0, 2))),
('logisticregression',
LogisticRegression())]),
Pipeline(steps=[('columnselector',
ColumnSelector(cols=(1, 2,
3))),
('logisticregression',
LogisticRegression())])],
meta_classifier=LogisticRegression(), random_state=42)
from sklearn.model_selection import cross_val_score cross_val_score(sclf,X,y,cv=5)
array([0.96666667, 0.96666667, 0.9 , 0.96666667, 1. ])