preface
In order to record my learning process, I have roughly sorted out the analysis process. The tool is to use Jupiter notebook, which I prefer, and then export it into md format and send it to csdn to share with you;
This is only a simple analysis process, so it is relatively simple. If there is something wrong, I hope you can include it more. You are also welcome to exchange opinions.
Table of Contents
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib
matplotlib.rcParams['font.family'] = 'STSong'
import seaborn as sns
from sklearn.ensemble import RandomForestClassifier
from sklearn.preprocessing import OneHotEncoder, LabelEncoder, StandardScaler
import warnings
warnings.filterwarnings('ignore')
data fetch
df_train = pd.read_csv('../titanic_dir/titanic_data/train.csv')
df_test = pd.read_csv('../titanic_dir/titanic_data/test.csv')
df_all = pd.concat([df_train, df_test])
df_all.reset_index(drop=True,inplace=True)
print('df_all'+'*'*20)
print(df_all.columns)
print('df_train'+'*'*20)
print(df_train.columns)
print('df_test'+'*'*20)
print(df_test.columns)
df_all********************
Index(['PassengerId', 'Survived', 'Pclass', 'Name', 'Sex', 'Age', 'SibSp',
'Parch', 'Ticket', 'Fare', 'Cabin', 'Embarked'],
dtype='object')
df_train********************
Index(['PassengerId', 'Survived', 'Pclass', 'Name', 'Sex', 'Age', 'SibSp',
'Parch', 'Ticket', 'Fare', 'Cabin', 'Embarked'],
dtype='object')
df_test********************
Index(['PassengerId', 'Pclass', 'Name', 'Sex', 'Age', 'SibSp', 'Parch',
'Ticket', 'Fare', 'Cabin', 'Embarked'],
dtype='object')
df_all
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0.0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| 1 | 2 | 1.0 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 2 | 3 | 1.0 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S |
| 3 | 4 | 1.0 | 1 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | female | 35.0 | 1 | 0 | 113803 | 53.1000 | C123 | S |
| 4 | 5 | 0.0 | 3 | Allen, Mr. William Henry | male | 35.0 | 0 | 0 | 373450 | 8.0500 | NaN | S |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1304 | 1305 | NaN | 3 | Spector, Mr. Woolf | male | NaN | 0 | 0 | A.5. 3236 | 8.0500 | NaN | S |
| 1305 | 1306 | NaN | 1 | Oliva y Ocana, Dona. Fermina | female | 39.0 | 0 | 0 | PC 17758 | 108.9000 | C105 | C |
| 1306 | 1307 | NaN | 3 | Saether, Mr. Simon Sivertsen | male | 38.5 | 0 | 0 | SOTON/O.Q. 3101262 | 7.2500 | NaN | S |
| 1307 | 1308 | NaN | 3 | Ware, Mr. Frederick | male | NaN | 0 | 0 | 359309 | 8.0500 | NaN | S |
| 1308 | 1309 | NaN | 3 | Peter, Master. Michael J | male | NaN | 1 | 1 | 2668 | 22.3583 | NaN | C |
1309 rows × 12 columns
df_all.info()
<class 'pandas.core.frame.DataFrame'> RangeIndex: 1309 entries, 0 to 1308 Data columns (total 12 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 PassengerId 1309 non-null int64 1 Survived 891 non-null float64 2 Pclass 1309 non-null int64 3 Name 1309 non-null object 4 Sex 1309 non-null object 5 Age 1046 non-null float64 6 SibSp 1309 non-null int64 7 Parch 1309 non-null int64 8 Ticket 1309 non-null object 9 Fare 1308 non-null float64 10 Cabin 295 non-null object 11 Embarked 1307 non-null object dtypes: float64(3), int64(4), object(5) memory usage: 122.8+ KB
The missing rate of Age and Cabin data is large;
Embarked, the deletion rate of far is very small
feature correlation
Pearson correlation formula
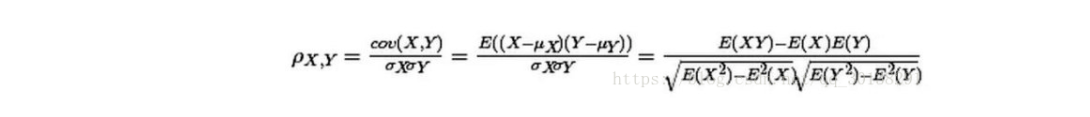
df_all.corr('pearson')
| PassengerId | Survived | Pclass | Age | SibSp | Parch | Fare | |
|---|---|---|---|---|---|---|---|
| PassengerId | 1.000000 | -0.005007 | -0.038354 | 0.028814 | -0.055224 | 0.008942 | 0.031428 |
| Survived | -0.005007 | 1.000000 | -0.338481 | -0.077221 | -0.035322 | 0.081629 | 0.257307 |
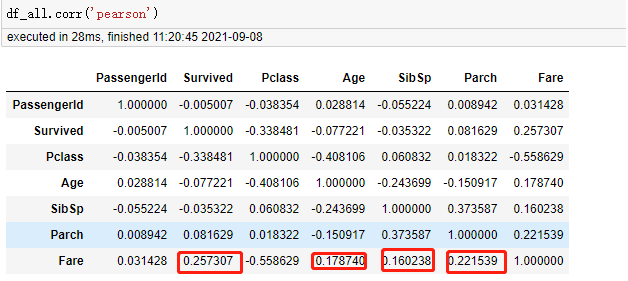
| Pclass | -0.038354 | -0.338481 | 1.000000 | -0.408106 | 0.060832 | 0.018322 | -0.558629 |
| Age | 0.028814 | -0.077221 | -0.408106 | 1.000000 | -0.243699 | -0.150917 | 0.178740 |
| SibSp | -0.055224 | -0.035322 | 0.060832 | -0.243699 | 1.000000 | 0.373587 | 0.160238 |
| Parch | 0.008942 | 0.081629 | 0.018322 | -0.150917 | 0.373587 | 1.000000 | 0.221539 |
| Fare | 0.031428 | 0.257307 | -0.558629 | 0.178740 | 0.160238 | 0.221539 | 1.000000 |
Data filling
df_all
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0.0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| 1 | 2 | 1.0 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 2 | 3 | 1.0 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S |
| 3 | 4 | 1.0 | 1 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | female | 35.0 | 1 | 0 | 113803 | 53.1000 | C123 | S |
| 4 | 5 | 0.0 | 3 | Allen, Mr. William Henry | male | 35.0 | 0 | 0 | 373450 | 8.0500 | NaN | S |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1304 | 1305 | NaN | 3 | Spector, Mr. Woolf | male | NaN | 0 | 0 | A.5. 3236 | 8.0500 | NaN | S |
| 1305 | 1306 | NaN | 1 | Oliva y Ocana, Dona. Fermina | female | 39.0 | 0 | 0 | PC 17758 | 108.9000 | C105 | C |
| 1306 | 1307 | NaN | 3 | Saether, Mr. Simon Sivertsen | male | 38.5 | 0 | 0 | SOTON/O.Q. 3101262 | 7.2500 | NaN | S |
| 1307 | 1308 | NaN | 3 | Ware, Mr. Frederick | male | NaN | 0 | 0 | 359309 | 8.0500 | NaN | S |
| 1308 | 1309 | NaN | 3 | Peter, Master. Michael J | male | NaN | 1 | 1 | 2668 | 22.3583 | NaN | C |
1309 rows × 12 columns
Fare
df_all[df_all['Fare'].isnull()]
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1043 | 1044 | NaN | 3 | Storey, Mr. Thomas | male | 60.5 | 0 | 0 | 3701 | NaN | NaN | S |
There is only one missing data in Fare, and it comes from the test verification data, so the observed is empty. The correlation between Age, sibsp and parch and Fare is greater than 0.1, and these three values in this line of data are not empty, but the Age here is a continuous value, which is difficult to group. Considering that there is only one missing data, Age is discarded because gender is generally considered, So finally, Sex is also added, so the median after grouping the three attributes SibSp,Parch and Sex - groupby is filled here
df_all.groupby(['Parch', 'SibSp','Sex'])['Fare'].median() # df_all.groupby(['Parch', 'Age', 'SibSp','Sex'])['Fare'].median().to_clipboard()
Parch SibSp Sex
0 0 female 10.50000
male 8.05000
1 female 26.00000
male 26.00000
2 female 23.25000
male 23.25000
3 female 18.42500
male 18.00000
1 0 female 39.40000
male 33.00000
1 female 26.00000
male 23.00000
2 female 23.00000
male 25.25000
3 female 25.46670
male 21.55000
4 male 34.40625
2 0 female 22.35830
male 30.75000
1 female 41.57920
male 41.57920
2 female 148.37500
male 148.37500
3 female 263.00000
male 27.90000
4 female 31.27500
male 31.38750
5 female 46.90000
male 46.90000
8 female 69.55000
male 69.55000
3 0 female 29.12915
1 female 34.37500
male 148.37500
2 female 18.75000
4 0 female 23.27085
1 female 145.45000
male 145.45000
5 0 female 34.40625
1 female 31.33125
male 31.33125
6 1 female 46.90000
male 46.90000
9 1 female 69.55000
male 69.55000
Name: Fare, dtype: float64
Parch: 0, sibsp: 0, sex: male: 8.05000
df_all['Fare'] = df_all['Fare'].fillna(8.05000)
df_all[df_all['Fare'].isnull()]
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked |
|---|
Age
263 rows missing Age
According to the Pearson correlation table, there are only Fare and Age related in all digital fields
Age, embarked and cabin can be considered for subsequent optimization
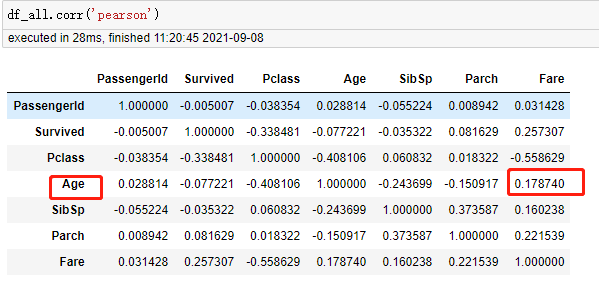
df_all['Age'].isnull().value_counts()
False 1046 True 263 Name: Age, dtype: int64
Observe the Age field and Fare
Take a look at the data distribution of Fare
plt.figure(figsize=(15,10)) plt.hist(df_all['Fare']) plt.show()
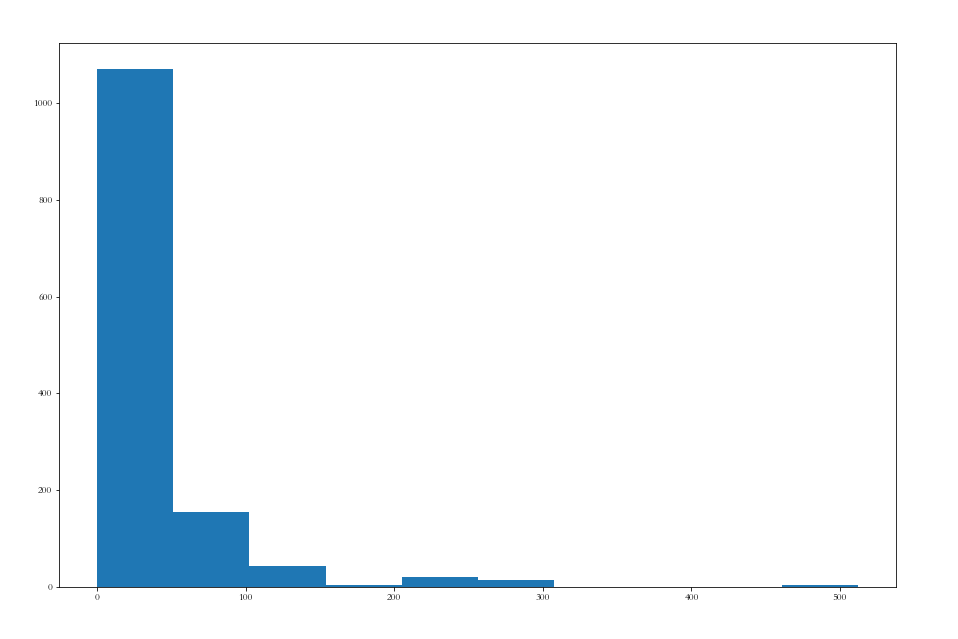
Most fare s are in the range of 0-50
# Isometric segmentation df_all['Fare_cut'] = pd.qcut(df_all['Fare'],10) plt.figure(figsize=(15,10)) df_all['Fare_cut'].value_counts().plot(kind='bar',rot=30)
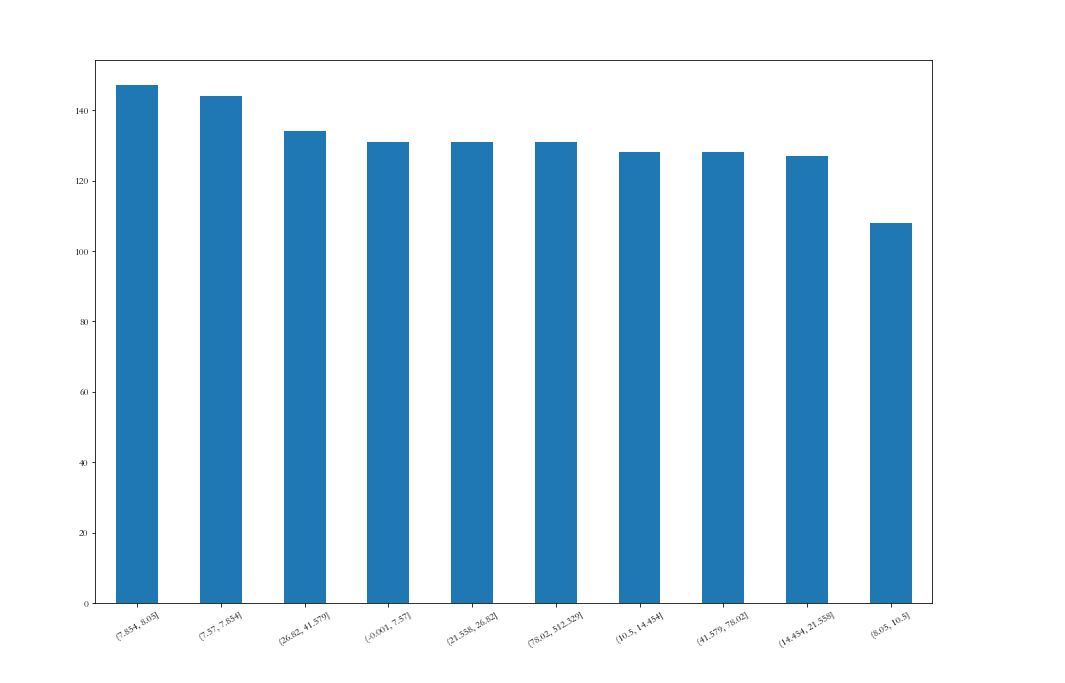
<AxesSubplot:>
Far is equally divided into 10 intervals
df_all.groupby(['Fare_cut','Sex']).agg({'Age':'median'})
| Age | ||
|---|---|---|
| Fare_cut | Sex | |
| (-0.001, 7.57] | female | 20.25 |
| male | 25.00 | |
| (7.57, 7.854] | female | 22.00 |
| male | 25.00 | |
| (7.854, 8.05] | female | 24.00 |
| male | 28.00 | |
| (8.05, 10.5] | female | 22.50 |
| male | 24.50 | |
| (10.5, 14.454] | female | 27.00 |
| male | 30.00 | |
| (14.454, 21.558] | female | 24.00 |
| male | 26.00 | |
| (21.558, 26.82] | female | 29.00 |
| male | 36.00 | |
| (26.82, 41.579] | female | 24.00 |
| male | 29.00 | |
| (41.579, 78.02] | female | 35.00 |
| male | 32.00 | |
| (78.02, 512.329] | female | 35.00 |
| male | 37.00 |
Group by. Transform after grouping, fill in with the median Age in the group (transform is really easy to use!!! Readers are recommended to learn more about Baidu)
df_all['Age'] = df_all.groupby(['Fare_cut','Sex'])['Age'].transform(lambda x:x.fillna(x.median()))
df_all['Age'].isnull().value_counts()
False 1309 Name: Age, dtype: int64
The difference between apply and transform in groupby
list(df_all.groupby(['Fare_cut','Sex']))
[((Interval(-0.001, 7.57, closed='right'), 'female'),
PassengerId Survived Pclass \
19 20 1.0 3
235 236 0.0 3
367 368 1.0 3
376 377 1.0 3
649 650 1.0 3
654 655 0.0 3
780 781 1.0 3
786 787 1.0 3
875 876 1.0 3
892 893 NaN 3
899 900 NaN 3
910 911 NaN 3
1004 1005 NaN 3
1182 1183 NaN 3
1238 1239 NaN 3
The data operated by apply in group by can be series or dataframe
The data operated by the transform in groupby can only be series
The following apply groups directly
df_all.groupby(['Fare_cut','Sex'])['Age','Fare'].apply(lambda x:x.count())
| Age | Fare | ||
|---|---|---|---|
| Fare_cut | Sex | ||
| (-0.001, 7.57] | female | 15 | 15 |
| male | 116 | 116 | |
| (7.57, 7.854] | female | 50 | 50 |
| male | 94 | 94 | |
| (7.854, 8.05] | female | 22 | 22 |
| male | 125 | 125 | |
| (8.05, 10.5] | female | 30 | 30 |
| male | 78 | 78 | |
| (10.5, 14.454] | female | 49 | 49 |
| male | 79 | 79 | |
| (14.454, 21.558] | female | 61 | 61 |
| male | 66 | 66 | |
| (21.558, 26.82] | female | 52 | 52 |
| male | 79 | 79 | |
| (26.82, 41.579] | female | 52 | 52 |
| male | 82 | 82 | |
| (41.579, 78.02] | female | 53 | 53 |
| male | 75 | 75 | |
| (78.02, 512.329] | female | 82 | 82 |
| male | 49 | 49 |
In the following apply, the X passed in is a dataframe. If you need to operate on a single column, you can use x.iloc[:,0]
df_all.groupby(['Fare_cut','Sex'])['Age','Fare'].apply(lambda x:x.iloc[:,0].count()+x.iloc[:,1].count()) # df_all.groupby(['Fare_cut','Sex'])['Age','Fare'].apply(lambda x:print(type(x)))
Fare_cut Sex
(-0.001, 7.57] female 30
male 232
(7.57, 7.854] female 100
male 188
(7.854, 8.05] female 44
male 250
(8.05, 10.5] female 60
male 156
(10.5, 14.454] female 98
male 158
(14.454, 21.558] female 122
male 132
(21.558, 26.82] female 104
male 158
(26.82, 41.579] female 104
male 164
(41.579, 78.02] female 106
male 150
(78.02, 512.329] female 164
male 98
dtype: int64
Embarked
df_all[df_all['Embarked'].isnull()]
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Fare_cut | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 61 | 62 | 1.0 | 1 | Icard, Miss. Amelie | female | 38.0 | 0 | 0 | 113572 | 80.0 | B28 | NaN | (78.02, 512.329] |
| 829 | 830 | 1.0 | 1 | Stone, Mrs. George Nelson (Martha Evelyn) | female | 62.0 | 0 | 0 | 113572 | 80.0 | B28 | NaN | (78.02, 512.329] |
df_all.groupby(['Fare_cut','Sex','Embarked'])['Embarked'].count()
Fare_cut Sex Embarked
(-0.001, 7.57] female C 7
Q 3
S 5
male C 42
Q 4
S 70
(7.57, 7.854] female C 0
Q 34
S 16
male C 0
Q 38
S 56
(7.854, 8.05] female C 0
Q 8
S 14
male C 6
Q 2
S 117
(8.05, 10.5] female C 1
Q 1
S 28
male C 3
Q 2
S 73
(10.5, 14.454] female C 12
Q 2
S 35
male C 11
Q 4
S 64
(14.454, 21.558] female C 13
Q 6
S 42
male C 15
Q 4
S 47
(21.558, 26.82] female C 3
Q 3
S 46
male C 8
Q 3
S 68
(26.82, 41.579] female C 15
Q 1
S 36
male C 27
Q 5
S 50
(41.579, 78.02] female C 20
Q 0
S 33
male C 17
Q 0
S 58
(78.02, 512.329] female C 42
Q 2
S 36
male C 28
Q 1
S 20
Name: Embarked, dtype: int64
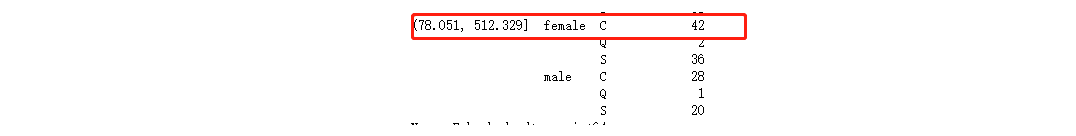
The missing two lines are both women and the cost is 80 yuan
Here, use the corresponding:
The ticket price is between (78.051, 512.329]
Among women, the largest number is C
# Filling the missing values in Embarked with S
df_all['Embarked'] = df_all['Embarked'].fillna('C')
df_all[df_all['Embarked'].isnull()]
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Fare_cut |
|---|
Cabin
df_all['Cabin'].isnull().value_counts()
True 1014 False 295 Name: Cabin, dtype: int64
The bin attribute has many missing values. Here, M is used to fill in the missing values temporarily, and the others are replaced by the initial letter
df_all['Cabin'] = df_all['Cabin'].fillna('M')
# The missing value is replaced by M
df_all['Cabin_new'] = df_all['Cabin'].apply(lambda x:x[0])
df_all['Cabin_new'].value_counts()
M 1014 C 94 B 65 D 46 E 41 A 22 F 21 G 5 T 1 Name: Cabin_new, dtype: int64
End of missing value supplement
df_all.info()
<class 'pandas.core.frame.DataFrame'> RangeIndex: 1309 entries, 0 to 1308 Data columns (total 14 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 PassengerId 1309 non-null int64 1 Survived 891 non-null float64 2 Pclass 1309 non-null int64 3 Name 1309 non-null object 4 Sex 1309 non-null object 5 Age 1309 non-null float64 6 SibSp 1309 non-null int64 7 Parch 1309 non-null int64 8 Ticket 1309 non-null object 9 Fare 1309 non-null float64 10 Cabin 1309 non-null object 11 Embarked 1309 non-null object 12 Fare_cut 1309 non-null category 13 Cabin_new 1309 non-null object dtypes: category(1), float64(3), int64(4), object(6) memory usage: 135.1+ KB
df_all_copy = df_all.copy() # Data backup
Rescued analysis
df_all
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Fare_cut | Cabin_new | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0.0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | M | S | (-0.001, 7.57] | M |
| 1 | 2 | 1.0 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C | (41.579, 78.02] | C |
| 2 | 3 | 1.0 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | M | S | (7.854, 8.05] | M |
| 3 | 4 | 1.0 | 1 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | female | 35.0 | 1 | 0 | 113803 | 53.1000 | C123 | S | (41.579, 78.02] | C |
| 4 | 5 | 0.0 | 3 | Allen, Mr. William Henry | male | 35.0 | 0 | 0 | 373450 | 8.0500 | M | S | (7.854, 8.05] | M |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1304 | 1305 | NaN | 3 | Spector, Mr. Woolf | male | 28.0 | 0 | 0 | A.5. 3236 | 8.0500 | M | S | (7.854, 8.05] | M |
| 1305 | 1306 | NaN | 1 | Oliva y Ocana, Dona. Fermina | female | 39.0 | 0 | 0 | PC 17758 | 108.9000 | C105 | C | (78.02, 512.329] | C |
| 1306 | 1307 | NaN | 3 | Saether, Mr. Simon Sivertsen | male | 38.5 | 0 | 0 | SOTON/O.Q. 3101262 | 7.2500 | M | S | (-0.001, 7.57] | M |
| 1307 | 1308 | NaN | 3 | Ware, Mr. Frederick | male | 28.0 | 0 | 0 | 359309 | 8.0500 | M | S | (7.854, 8.05] | M |
| 1308 | 1309 | NaN | 3 | Peter, Master. Michael J | male | 36.0 | 1 | 1 | 2668 | 22.3583 | M | C | (21.558, 26.82] | M |
1309 rows × 14 columns
survived_sum = df_train['Survived'].value_counts().sum() df_train['Survived'].value_counts()/ survived_sum
0 0.616162 1 0.383838 Name: Survived, dtype: float64
(df_train['Survived'].value_counts()/ survived_sum).plot(kind='bar')
<AxesSubplot:>
The probability of being rescued in training concentration is about 38%
Feature Engineering
Fare
del df_all['Fare_cut'] df_all[:2]
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Cabin_new | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0.0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | M | S | M |
| 1 | 2 | 1.0 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C | C |
When filling in the missing value for the Age attribute, the far field is divided into 10 intervals, and a new attribute far_cut is created, which is deleted here
Cabin
df_all['Cabin'] = df_all['Cabin_new'] del df_all['Cabin_new']
df_all[:2]
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0.0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | M | S |
| 1 | 2 | 1.0 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C | C |
Replace bin with the value of the newly built attribute bin_new
Age
Not yet
New feature --- family_size
df_all['Family_Size'] = df_all['SibSp'] + df_all['Parch'] + 1
Add 1 to SibSp and Parch to obtain the value of Family_Size
df_all[:2]
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Family_Size | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0.0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | M | S | 2 |
| 1 | 2 | 1.0 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C | C | 2 |
New feature --- title
df_all['Name'].str.split(', ', expand=True)
| 0 | 1 | |
|---|---|---|
| 0 | Braund | Mr. Owen Harris |
| 1 | Cumings | Mrs. John Bradley (Florence Briggs Thayer) |
| 2 | Heikkinen | Miss. Laina |
| 3 | Futrelle | Mrs. Jacques Heath (Lily May Peel) |
| 4 | Allen | Mr. William Henry |
| ... | ... | ... |
| 1304 | Spector | Mr. Woolf |
| 1305 | Oliva y Ocana | Dona. Fermina |
| 1306 | Saether | Mr. Simon Sivertsen |
| 1307 | Ware | Mr. Frederick |
| 1308 | Peter | Master. Michael J |
1309 rows × 2 columns
Extract the prefix in the Name field
df_all['Title'] = df_all['Name'].str.split(', ', expand=True)[1].str.split('.', expand=True)[0]
df_all['Title'].value_counts()
Mr 757 Miss 260 Mrs 197 Master 61 Dr 8 Rev 8 Col 4 Major 2 Mlle 2 Ms 2 Lady 1 Capt 1 Jonkheer 1 Mme 1 Dona 1 Sir 1 the Countess 1 Don 1 Name: Title, dtype: int64
plt.subplots(figsize=(20, 10))
sns.barplot(x=df_all['Title'].value_counts().index, y=df_all['Title'].value_counts().values)
plt.title('Number of prefixes')
plt.show()
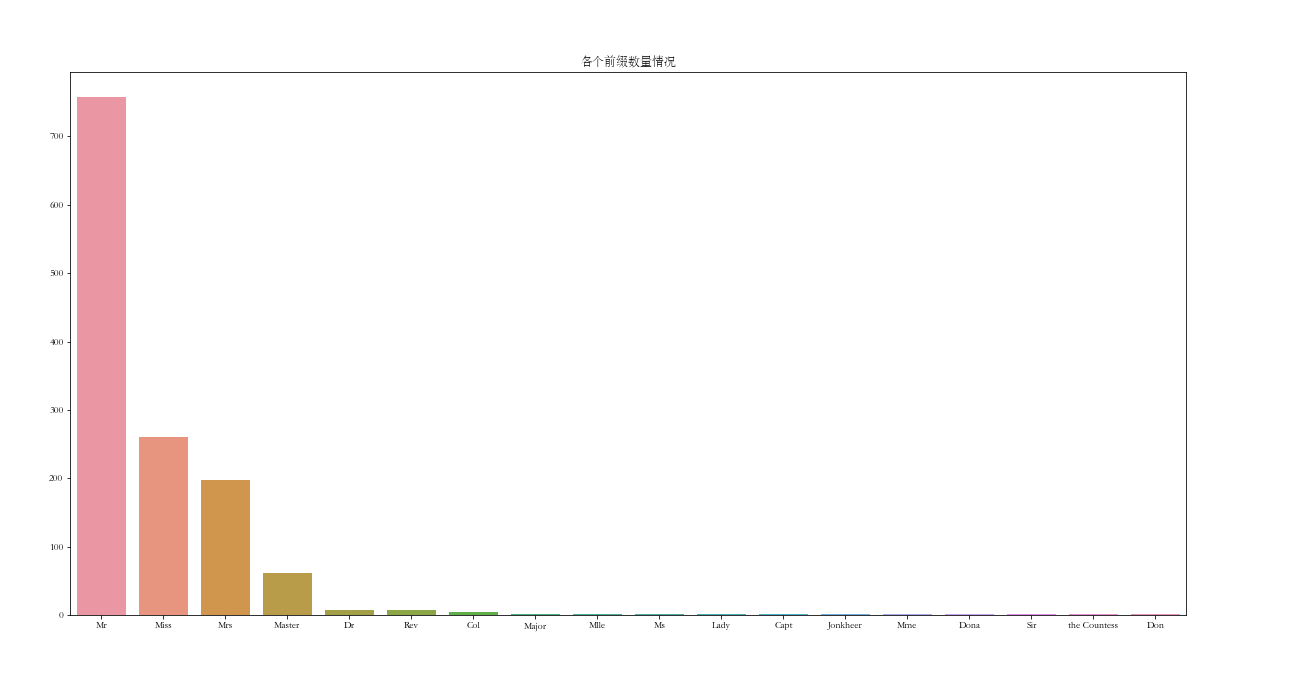
Here, I put the common Mr, Miss, Mrs and Ms into one category, and the others into one category
df_all['Title'].replace(['Mr','Miss','Mrs','Ms'],'cate1',inplace=True) df_all['Title'] = df_all['Title'].apply(lambda x:'cate1' if x=='cate1' else 'cate2')
df_all['Title'].value_counts()
cate1 1216 cate2 93 Name: Title, dtype: int64
cate1 stands for Mr, Miss, Mrs and Ms, and cate2 stands for others
df_all[:3]
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Family_Size | Title | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0.0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | M | S | 2 | cate1 |
| 1 | 2 | 1.0 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C | C | 2 | cate1 |
| 2 | 3 | 1.0 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | M | S | 1 | cate1 |
Delete feature --- passengerid, name, ticket
df_all.drop(columns=['PassengerId','Name','Ticket'],inplace=True) df_all[:3]
| Survived | Pclass | Sex | Age | SibSp | Parch | Fare | Cabin | Embarked | Family_Size | Title | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0 | 3 | male | 22.0 | 1 | 0 | 7.2500 | M | S | 2 | cate1 |
| 1 | 1.0 | 1 | female | 38.0 | 1 | 0 | 71.2833 | C | C | 2 | cate1 |
| 2 | 1.0 | 3 | female | 26.0 | 0 | 0 | 7.9250 | M | S | 1 | cate1 |
Classification feature coding
Nominal variable and distance variable are suitable for OneHotEncoder
Distance variable, suitable for LabelEncoder

df_all
| Survived | Pclass | Sex | Age | SibSp | Parch | Fare | Cabin | Embarked | Family_Size | Title | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0 | 3 | male | 22.0 | 1 | 0 | 7.2500 | M | S | 2 | cate1 |
| 1 | 1.0 | 1 | female | 38.0 | 1 | 0 | 71.2833 | C | C | 2 | cate1 |
| 2 | 1.0 | 3 | female | 26.0 | 0 | 0 | 7.9250 | M | S | 1 | cate1 |
| 3 | 1.0 | 1 | female | 35.0 | 1 | 0 | 53.1000 | C | S | 2 | cate1 |
| 4 | 0.0 | 3 | male | 35.0 | 0 | 0 | 8.0500 | M | S | 1 | cate1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1304 | NaN | 3 | male | 28.0 | 0 | 0 | 8.0500 | M | S | 1 | cate1 |
| 1305 | NaN | 1 | female | 39.0 | 0 | 0 | 108.9000 | C | C | 1 | cate2 |
| 1306 | NaN | 3 | male | 38.5 | 0 | 0 | 7.2500 | M | S | 1 | cate1 |
| 1307 | NaN | 3 | male | 28.0 | 0 | 0 | 8.0500 | M | S | 1 | cate1 |
| 1308 | NaN | 3 | male | 36.0 | 1 | 1 | 22.3583 | M | C | 3 | cate2 |
1309 rows × 11 columns
Sex,Cabin,Embarked,Title
The four feature s to be encoded here should be encoded with onehot
OneHotEncoder
cat_features_list = ['Sex', 'Cabin', 'Embarked', 'Title']
df_all_encode = pd.DataFrame()
for feature in cat_features_list:
data_encode = OneHotEncoder().fit_transform(df_all[feature].values.reshape(-1, 1)).toarray()
value_count = df_all[feature].unique().size
new_columns = ['{}_{}'.format(feature,i) for i in range(1,value_count+1)]
print(new_columns)
df_encode = pd.DataFrame(data_encode,columns=new_columns)
# print(df_encode)
df_all_encode = pd.concat([df_all_encode,df_encode],axis=1)
['Sex_1', 'Sex_2'] ['Cabin_1', 'Cabin_2', 'Cabin_3', 'Cabin_4', 'Cabin_5', 'Cabin_6', 'Cabin_7', 'Cabin_8', 'Cabin_9'] ['Embarked_1', 'Embarked_2', 'Embarked_3'] ['Title_1', 'Title_2']
The data encoded by OneHotEncoder is as follows
df_all_encode
| Sex_1 | Sex_2 | Cabin_1 | Cabin_2 | Cabin_3 | Cabin_4 | Cabin_5 | Cabin_6 | Cabin_7 | Cabin_8 | Cabin_9 | Embarked_1 | Embarked_2 | Embarked_3 | Title_1 | Title_2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 |
| 1 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 2 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 |
| 3 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 |
| 4 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1304 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 |
| 1305 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 1306 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 |
| 1307 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 |
| 1308 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 |
1309 rows × 16 columns
Splice the data generated by OneHotEncoder encoding and df_all, and delete the four feature s before encoding: ['Sex', 'Cabin', 'embanked', 'Title']
df_all = pd.concat([df_all,df_all_encode],axis=1) df_all.drop(columns=['Sex','Cabin','Embarked','Title'],inplace=True)
df_all[:3]
| Survived | Pclass | Age | SibSp | Parch | Fare | Family_Size | Sex_1 | Sex_2 | Cabin_1 | ... | Cabin_5 | Cabin_6 | Cabin_7 | Cabin_8 | Cabin_9 | Embarked_1 | Embarked_2 | Embarked_3 | Title_1 | Title_2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0 | 3 | 22.0 | 1 | 0 | 7.2500 | 2 | 0.0 | 1.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 |
| 1 | 1.0 | 1 | 38.0 | 1 | 0 | 71.2833 | 2 | 1.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 2 | 1.0 | 3 | 26.0 | 0 | 0 | 7.9250 | 1 | 1.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 |
3 rows × 23 columns
df_all.info()
<class 'pandas.core.frame.DataFrame'> RangeIndex: 1309 entries, 0 to 1308 Data columns (total 23 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 Survived 891 non-null float64 1 Pclass 1309 non-null int64 2 Age 1309 non-null float64 3 SibSp 1309 non-null int64 4 Parch 1309 non-null int64 5 Fare 1309 non-null float64 6 Family_Size 1309 non-null int64 7 Sex_1 1309 non-null float64 8 Sex_2 1309 non-null float64 9 Cabin_1 1309 non-null float64 10 Cabin_2 1309 non-null float64 11 Cabin_3 1309 non-null float64 12 Cabin_4 1309 non-null float64 13 Cabin_5 1309 non-null float64 14 Cabin_6 1309 non-null float64 15 Cabin_7 1309 non-null float64 16 Cabin_8 1309 non-null float64 17 Cabin_9 1309 non-null float64 18 Embarked_1 1309 non-null float64 19 Embarked_2 1309 non-null float64 20 Embarked_3 1309 non-null float64 21 Title_1 1309 non-null float64 22 Title_2 1309 non-null float64 dtypes: float64(19), int64(4) memory usage: 235.3 KB
Data segmentation
df_train = df_all.loc[:890] df_test = df_all.loc[891:] del df_test['Survived'] y_train = df_train['Survived'].values del df_train['Survived']
df_train.shape
(891, 22)
df_test.shape
(418, 22)
y_train.shape
(891,)
Standardization
x_train = StandardScaler().fit_transform(df_train)
x_test = StandardScaler().fit_transform(df_test)
print('x_train shape: {}'.format(x_train.shape))
print('y_train shape: {}'.format(y_train.shape))
print('x_test shape: {}'.format(x_test.shape))
x_train shape: (891, 22) y_train shape: (891,) x_test shape: (418, 22)
model
Decision tree
df_result = pd.DataFrame()
from sklearn import tree decision_tree_model = tree.DecisionTreeClassifier() decision_tree_model.fit(x_train,y_train) y_predict_with_dtree = decision_tree_model.predict(x_test) df_result['y_predict_with_dtree'] = y_predict_with_dtree df_result
| y_predict_with_dtree | |
|---|---|
| 0 | 0.0 |
| 1 | 0.0 |
| 2 | 1.0 |
| 3 | 1.0 |
| 4 | 1.0 |
| ... | ... |
| 413 | 0.0 |
| 414 | 1.0 |
| 415 | 0.0 |
| 416 | 0.0 |
| 417 | 1.0 |
418 rows × 1 columns
logistic regression
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression().fit(x_train,y_train) y_predict_with_logisticReg = lr.predict(x_test) df_result['y_predict_with_logisticReg'] = y_predict_with_logisticReg df_result
| y_predict_with_dtree | y_predict_with_logisticReg | |
|---|---|---|
| 0 | 0.0 | 0.0 |
| 1 | 0.0 | 0.0 |
| 2 | 1.0 | 0.0 |
| 3 | 1.0 | 0.0 |
| 4 | 1.0 | 1.0 |
| ... | ... | ... |
| 413 | 0.0 | 0.0 |
| 414 | 1.0 | 1.0 |
| 415 | 0.0 | 0.0 |
| 416 | 0.0 | 0.0 |
| 417 | 1.0 | 0.0 |
418 rows × 2 columns
Support vector machine
from sklearn import svm
svm_model = svm.SVC() svm_model.fit(x_train,y_train) y_predict_with_svm = svm_model.predict(x_test) df_result['y_predict_with_svm'] = y_predict_with_svm df_result
| y_predict_with_dtree | y_predict_with_logisticReg | y_predict_with_svm | |
|---|---|---|---|
| 0 | 0.0 | 0.0 | 0.0 |
| 1 | 0.0 | 0.0 | 1.0 |
| 2 | 1.0 | 0.0 | 0.0 |
| 3 | 1.0 | 0.0 | 0.0 |
| 4 | 1.0 | 1.0 | 0.0 |
| ... | ... | ... | ... |
| 413 | 0.0 | 0.0 | 0.0 |
| 414 | 1.0 | 1.0 | 1.0 |
| 415 | 0.0 | 0.0 | 0.0 |
| 416 | 0.0 | 0.0 | 0.0 |
| 417 | 1.0 | 0.0 | 1.0 |
418 rows × 3 columns
KNN
from sklearn import neighbors
knnmodel = neighbors.KNeighborsClassifier(n_neighbors=2) #The n_neighbors parameter is the number of classifications knnmodel.fit(x_train,y_train) y_predict_with_knn = knnmodel.predict(x_test) df_result['y_predict_with_knn'] = y_predict_with_knn df_result
| y_predict_with_dtree | y_predict_with_logisticReg | y_predict_with_svm | y_predict_with_knn | |
|---|---|---|---|---|
| 0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 1 | 0.0 | 0.0 | 1.0 | 0.0 |
| 2 | 1.0 | 0.0 | 0.0 | 0.0 |
| 3 | 1.0 | 0.0 | 0.0 | 0.0 |
| 4 | 1.0 | 1.0 | 0.0 | 0.0 |
| ... | ... | ... | ... | ... |
| 413 | 0.0 | 0.0 | 0.0 | 0.0 |
| 414 | 1.0 | 1.0 | 1.0 | 1.0 |
| 415 | 0.0 | 0.0 | 0.0 | 0.0 |
| 416 | 0.0 | 0.0 | 0.0 | 0.0 |
| 417 | 1.0 | 0.0 | 1.0 | 1.0 |
418 rows × 4 columns
Random forest
from sklearn.ensemble import RandomForestClassifier
model_randomforest = RandomForestClassifier().fit(x_train,y_train) y_predict_with_random_forest = model_randomforest.predict(x_test) df_result['y_predict_with_random_forest'] = y_predict_with_random_forest df_result
| y_predict_with_dtree | y_predict_with_logisticReg | y_predict_with_svm | y_predict_with_knn | y_predict_with_random_forest | |
|---|---|---|---|---|---|
| 0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 1 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 2 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 3 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 4 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| ... | ... | ... | ... | ... | ... |
| 413 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 414 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| 415 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 416 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 417 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 |
418 rows × 5 columns
Result verification
Here, in order to quickly obtain the accuracy of my prediction data locally, I obtained the prediction results with 100% accuracy from the records submitted by other gods of kaggle as the verification data;
Therefore, the accuracy of the above five models is as follows, and the random forest score is the highest: 0.7871.
df_check = pd.read_csv(r'../titanic_dir/titanic_data/correct_submission_titanic.csv')
df_check = df_check['Survived']
# df_check
for column in df_result:
df_concat = pd.concat([df_result[column],df_check],axis=1)
df_concat['predict_tag'] = df_concat.apply(lambda x: 1 if x[0]==x[1] else 0,axis=1)
right_rate = df_concat['predict_tag'].sum()/df_concat['predict_tag'].count()
print(column,'The accuracy is:')
print(np.round(right_rate,4))
y_predict_with_dtree The accuracy is: 0.7057 y_predict_with_logisticReg The accuracy is: 0.7703 y_predict_with_svm The accuracy is: 0.7656 y_predict_with_knn The accuracy is: 0.7656 y_predict_with_random_forest The accuracy is: 0.7871
Result regression optimization
1
When filling in the missing value, Bin: the Cabin has only been dealt with briefly, and there is no good analysis on how to deal with it. I feel that different positions of the Cabin still have a great impact on the probability of being rescued. Moreover, the proportion of missing values of this attribute is very large, and the method of missing value treatment should have a great impact on the results. When optimizing later, we should spend more time thinking about how to do it ;
2
When you call the model, you simply call the five models without selecting some parameters. During later optimization, you can filter the parameters for each model, and then predict. The results should be much better.