Polynomial logistic regression is to add high-order terms as features on the basis of logistic regression to realize the extraction of high-dimensional features
Model construction
Polynomial logistic regression model is composed of three sub models:
(1) Add polynomial feature
(2) Standardization
(3) Logistic regression
Add polynomial feature
Multiply each feature to obtain a new feature. For example, the original feature is \ ([x_0,x_1] \)
Quadratic polynomials are characterized by \ ([1,x_0,x_1,x_0^2,x_0x_1,x_1^2] \)
Cubic polynomials are characterized by \ ([1,x_0,x_1,x_0^2,x_0x_1,x_1^2,x_0^3,x_0^2x_1,x_0x_1^2,x_1^3] \)
def poly_transform(X):
XX=X.T
ret=[np.repeat(1.0,XX.shape[1]),XX[0],XX[1]]
for i in range(2,degree+1):
for j in range(0,i+1):
ret.append(XX[0]**(i-j)*XX[1]**j)
return np.array(ret).T
Super parameter setting:
degree=3
Standardization
Scale the sample to a mean value of 0, a standard deviation of 1, and a transformation formula of \ (\ Large x=\frac{x-\mu}{\sigma} \)
Pit point:
(1) The standard deviation may be 0. When converting, the denominator will be 0, which needs to be converted to a non-zero number
(2) You only need to fit the training data once and transform it according to the mean and standard deviation during training, otherwise there will be deviation
def scaler_fit(X):
global mean,scale
mean=X.mean(axis=0)
scale=X.std(axis=0)
scale[scale<np.finfo(scale.dtype).eps]=1.0
def scaler_transform(X):
return (X-mean)/scale
logistic regression
The core part of the model, the training process consists of three parts: forward propagation, loss calculation and back propagation:
Forward propagation
Calculate output probability from input data \ (p(x) \)
def sigmoid(x):
return 1/(1+exp(-x))
def forward(x):
return sigmoid(w@x.T)
In a sense \ (p(x) \) represents the probability that \ (x \) is judged as class 1
It is reasonable to add an offset term \ (b \), which may be better for some data
Calculate loss
Suppose that the probability of training data \ (x \) \ (p=p(x) \) has been obtained in the previous step
For a single training data \ ([p,y] \), the loss function is:
Using MBGD, multiple training data are obtained at one time. For a group of batch with m training data, the following are:
In order to prevent the weight of \ (w \) from expanding excessively (because the greater the absolute value of each weight of \ (w \), the closer the value of \ (p \) is to 0 or 1), L1 regular term is added. Therefore, the final loss function is:
def compute_loss(p,y):
return np.mean(-(y*log(p)+(1-y)*log(1-p)),axis=0)+k*np.linalg.norm(w,ord=1)
Back propagation + update parameters
Calculate the partial derivative of each weight of loss function \ (J \) to \ (w \)
For convenience of thinking, consider only the case of single data \ ([p,y] \):
So there
For m training data:
After adding regular items:
The form written as a vector is:
Update parameters while back propagating, i.e.:
\(\ alpha \) represents the learning rate
def backward(p,x,y):
global w
w-=learning_rate*np.mean((p-y).reshape(-1,1)*x,axis=0)+k*sign(w)
train
MBGD is used for training, and the learning rate is set as learning_rate, the weight of the regular item is k, and the training data scale of a batch is batch_size, the iteration round is epoch_num:
def logistic_fit(X,Y):
global w
XX=X.copy()
YY=Y.copy()
w=random.randn(XX[0].shape[0])*0.01
I=list(range(len(XX)))
LOSS=[]
for epoch in range(epoch_num):
random.shuffle(I)
XX=XX[I]
YY=YY[I]
for i in range(0,len(XX),batch_size):
x=XX[i:i+batch_size]
y=YY[i:i+batch_size]
p=forward(x)
loss=compute_loss(p,y)
LOSS.append(loss)
backward(p,x,y)
return LOSS
Super parameter setting:
learning_rate=0.9 k=0.005 batch_size=32 epoch_num=50
2, Generate training data
Generate training data sets \ (X \) and \ (Y \), where \ (X=\{x \} \), \ (Y=\{y \} \), \ (x=[x_0,F(x_0)] \), \ (x_0 ∈ [l,r] \), the label is \ (Y \), and the data set size is n
def generate_data(F,l,r,n,y):
x=np.linspace(l,r,n)
X=np.column_stack((x,F(x)))
Y=np.repeat(y,n)
return X,Y
Three, model training and prediction
train
Enter the training sample \ ((X,Y) \), update the parameters of scaler and logistic progression, and draw the loss curve with no return value
def train(X,Y):
XX=poly_transform(X)
scaler_fit(XX)
XX=scaler_transform(XX)
LOSS=logistic_fit(XX,Y)
plt.plot(list(range(len(LOSS))),LOSS,color='r')
plt.show()
forecast
Enter the prediction sample \ (X \) and return the prediction result \ (\ hat Y \)
def predict(X):
XX=scaler_transform(poly_transform(X))
return logistic_predict(XX)
Draw discrimination interface
Input prediction sample \ ((X,Y) \), scatter diagram, and discrimination interface
def plot_decision_boundary(X,Y):
global predY1
x0_min,x0_max=X[:,0].min()-1,X[:,0].max()+1
x1_min,x1_max=X[:,1].min()-1,X[:,1].max()+1
m=500
x0,x1=np.meshgrid(
np.linspace(x0_min,x0_max,m),
np.linspace(x1_min,x1_max,m)
)
XX=np.c_[x0.ravel(),x1.ravel()]
Y_pred=predict(XX)
Z=Y_pred.reshape(x0.shape)
plt.contourf(x0,x1,Z,cmap=plt.cm.Spectral)
plt.scatter(X[:,0],X[:,1],c=Y,cmap=plt.cm.Spectral)
plt.show()
4, Main program
random.seed(114514) data_size=200 X1,Y1=generate_data(lambda x:x**2+2*x-2+random.randn(data_size)*0.8,-3,1,data_size,0) X2,Y2=generate_data(lambda x:-x**2+2*x+2+random.randn(data_size)*0.8,-1,3,data_size,1) X=np.vstack((X1,X2)) Y=np.hstack((Y1,Y2)) train(X,Y) plot_decision_boundary(X,Y)
loss curve:
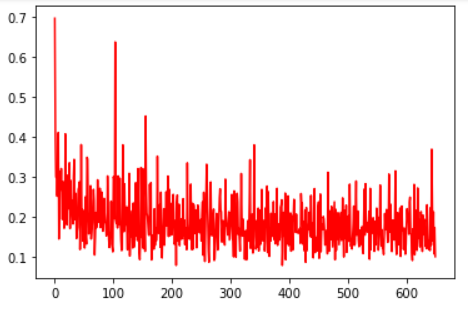
Discrimination interface:
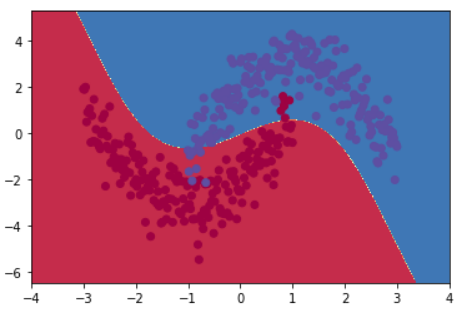
5, Complete code
Import dependent packagesmodel buildingimport numpy as np from numpy import exp,log,sign,random from matplotlib import pyplot as plt
# LogisticRegression
learning_rate=0.9
k=0.005
batch_size=32
epoch_num=50
def sigmoid(x):
return 1/(1+exp(-x))
def forward(x):
return sigmoid(w@x.T)
def compute_loss(p,y):
return np.mean(-(y*log(p)+(1-y)*log(1-p)),axis=0)+k*np.linalg.norm(w,ord=1)
def backward(p,x,y):
global w
w-=learning_rate*np.mean((p-y).reshape(-1,1)*x,axis=0)+k*sign(w)
def logistic_fit(X,Y):
global w
XX=X.copy()
YY=Y.copy()
w=random.randn(XX[0].shape[0])*0.01
I=list(range(len(XX)))
LOSS=[]
for epoch in range(epoch_num):
random.shuffle(I)
XX=XX[I]
YY=YY[I]
for i in range(0,len(XX),batch_size):
x=XX[i:i+batch_size]
y=YY[i:i+batch_size]
p=forward(x)
loss=compute_loss(p,y)
LOSS.append(loss)
backward(p,x,y)
return LOSS
def logistic_predict(X):
return np.where(forward(X)>0.5,1,0)
# StandardScaler
def scaler_fit(X):
global mean,scale
mean=X.mean(axis=0)
scale=X.std(axis=0)
scale[scale<np.finfo(scale.dtype).eps]=1.0
def scaler_transform(X):
return (X-mean)/scale
# PolynomialFeatures
degree=3
def poly_transform(X):
XX=X.T
ret=[np.repeat(1.0,XX.shape[1]),XX[0],XX[1]]
for i in range(2,degree+1):
for j in range(0,i+1):
ret.append(XX[0]**(i-j)*XX[1]**j)
return np.array(ret).T
def train(X,Y):
XX=poly_transform(X)
scaler_fit(XX)
XX=scaler_transform(XX)
LOSS=logistic_fit(XX,Y)
plt.plot(list(range(len(LOSS))),LOSS,color='r')
plt.show()
def predict(X):
XX=scaler_transform(poly_transform(X))
return logistic_predict(XX)
def plot_decision_boundary(X,Y):
global predY1
x0_min,x0_max=X[:,0].min()-1,X[:,0].max()+1
x1_min,x1_max=X[:,1].min()-1,X[:,1].max()+1
m=500
x0,x1=np.meshgrid(
np.linspace(x0_min,x0_max,m),
np.linspace(x1_min,x1_max,m)
)
XX=np.c_[x0.ravel(),x1.ravel()]
Y_pred=predict(XX)
Z=Y_pred.reshape(x0.shape)
plt.contourf(x0,x1,Z,cmap=plt.cm.Spectral)
plt.scatter(X[:,0],X[:,1],c=Y,cmap=plt.cm.Spectral)
plt.show()
def generate_data(F,l,r,n,y):
x=np.linspace(l,r,n)
X=np.column_stack((x,F(x)))
Y=np.repeat(y,n)
return X,Y
random.seed(114514) data_size=200 X1,Y1=generate_data(lambda x:x**2+2*x-2+random.randn(data_size)*0.8,-3,1,data_size,0) X2,Y2=generate_data(lambda x:-x**2+2*x+2+random.randn(data_size)*0.8,-1,3,data_size,1) X=np.vstack((X1,X2)) Y=np.hstack((Y1,Y2)) train(X,Y) plot_decision_boundary(X,Y)
reference material
- [1] Machine learning: logistic regression (using polynomial features)
- [2] Deep learning PyTorch minimalist tutorial