Problems caused by missing XGBoost values and their depth analysis
Author: Li Zhaojun, August 15, 2019 Article Links 3,969 words 8 minutes reading
1. Background
The XGBoost model, as a "killer" in machine learning, is widely used in data science contests and industries. XGBoost officially provides corresponding codes that can run on a variety of platforms and environments, such as XGBoost on Spark for distributed training.However, in the official implementation of XGBoost on Spark, there is a problem of instability caused by XGBoost missing values and sparse representation mechanisms.
It originated from feedback from students who used a machine learning platform in Meituan. The XGBoost model trained on the platform used the same model, the same test data, and the results calculated by local calls (Java engine) were inconsistent with those calculated by the platform (Spark engine).However, the student ran both engines locally (Python engine and Java engine) and the results were consistent.So would it be problematic to question the platform's XGBoost predictions?
The platform has many directional optimizations for XGBoost models, and there are no inconsistencies between local call (Java engine) and platform (Spark engine) results in XGBoost model tests.Moreover, the version running on the platform, and the version used locally by the students, all come from the official version of Dmlc. The underlying JNI calls should be the same code. In theory, the results should be identical, but in practice they are different.
From the test code given by the classmate, no problems were found:
//One row in the test result, 41 columns
double[] input = new double[]{1, 2, 5, 0, 0, 6.666666666666667, 31.14, 29.28, 0, 1.303333, 2.8555, 2.37, 701, 463, 3.989, 3.85, 14400.5, 15.79, 11.45, 0.915, 7.05, 5.5, 0.023333, 0.0365, 0.0275, 0.123333, 0.4645, 0.12, 15.082, 14.48, 0, 31.8425, 29.1, 7.7325, 3, 5.88, 1.08, 0, 0, 0, 32];
//Convert to float[]
float[] testInput = new float[input.length];
for(int i = 0, total = input.length; i < total; i++){
testInput[i] = new Double(input[i]).floatValue();
}
//Load Model
Booster booster = XGBoost.loadModel("${model}");
//Convert to DMatrix, one row, 41 columns
DMatrix testMat = new DMatrix(testInput, 1, 41);
//Call Model
float[][] predicts = booster.predict(testMat);
The result of executing the above code locally is 333.67892, whereas that on the platform is 328.1694030761719.
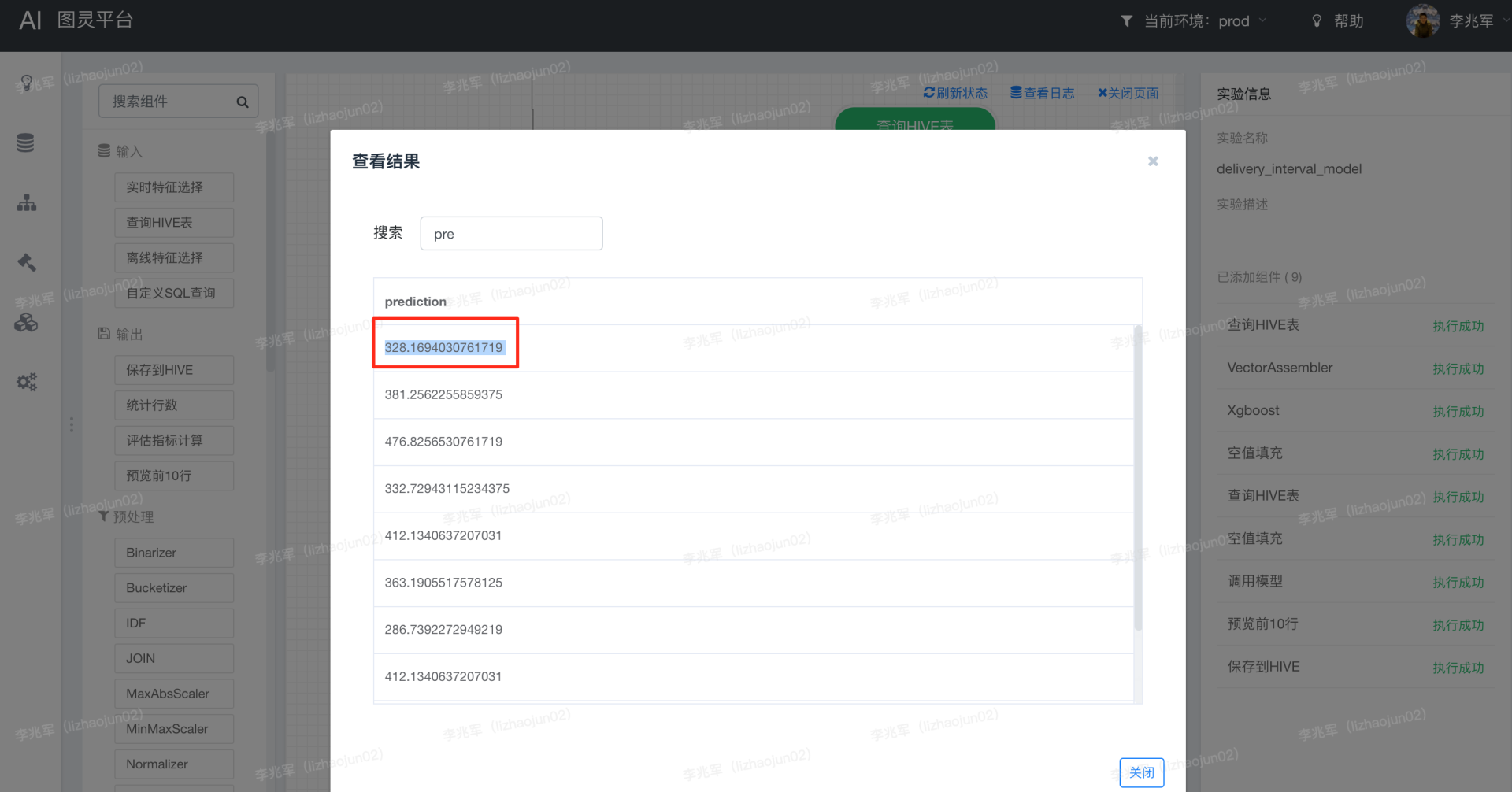
How can the results be different and where did the problem occur?
2. Process of checking inconsistent results
How to check?The first thing to think about is the direction of sorting out whether the field types entered in the two processes will be inconsistent.If the field types in the two inputs are inconsistent or the decimal precision is different, the difference in results is explanatory.After carefully analyzing the input of the model, I notice that there is a 6.6666666666667 in the array, is that why?
Each Debug carefully compares the input data and its field type on both sides and is identical.
This eliminates the problem of inconsistent field types and precision when dealing with both methods.
The second idea is that XGBoost on Spark provides two upper APIs, XGBoost Classifier and XGBoostRegressor, according to the function of the model. These two upper APIs add many hyperparameters and encapsulate many upper capabilities on the basis of JNI.Will it be that during these two encapsulations, some of the newly added hyperparameters have special handling of the input results, resulting in inconsistent results?
After communicating with the students who feedback this question, we know that the hyperparameters set in their Python code are exactly the same as the platform settings.Check the source code of XGBoostClassifier and XGBoostRegressor carefully, both of which do not have any special handling of the output results.
XGBoost on Spark hyperparameter encapsulation is again excluded.
Check the input of the model again, this time the idea is to check if there are any special values in the input of the model, for example, NaN, -1, 0, etc.Sure, there are several zeros in the input array, is it because of missing values?
Quickly find the source code of the two engines and find that they really do not handle the missing values in the same way!
Handling missing values in XGBoost4j
The processing of XGBoost4j missing values occurs during the construction of DMatrix, with 0.0f set as the missing value by default:
/**
* create DMatrix from dense matrix
*
* @param data data values
* @param nrow number of rows
* @param ncol number of columns
* @throws XGBoostError native error
*/
public DMatrix(float[] data, int nrow, int ncol) throws XGBoostError {
long[] out = new long[1];
//0.0f as missing value
XGBoostJNI.checkCall(XGBoostJNI.XGDMatrixCreateFromMat(data, nrow, ncol, 0.0f, out));
handle = out[0];
}
Handling missing values in XGBoost on Spark
xgboost on Spark uses NaN as the default missing value.
/**
* @return A tuple of the booster and the metrics used to build training summary
*/
@throws(classOf[XGBoostError])
def trainDistributed(
trainingDataIn: RDD[XGBLabeledPoint],
params: Map[String, Any],
round: Int,
nWorkers: Int,
obj: ObjectiveTrait = null,
eval: EvalTrait = null,
useExternalMemory: Boolean = false,
//NaN as missing value
missing: Float = Float.NaN,
hasGroup: Boolean = false): (Booster, Map[String, Array[Float]]) = {
//...
}
That is, when a local Java call constructs DMatrix, the default value of 0 is treated as the missing value if no missing value is set.In XGBoost on Spark, the default NaN is the missing value.The default missing values for the original Java engine are different from those for the XGBoost on Spark engine.However, when the platform and the classmate invoked, no missing values were set, which caused the inconsistent results between the two engines, because the missing values were inconsistent!
Modify the test code, set the missing value to NaN on the Java engine code, and execute it at 328.1694, which is exactly the same as the platform results.
//One row in the test result, 41 columns
double[] input = new double[]{1, 2, 5, 0, 0, 6.666666666666667, 31.14, 29.28, 0, 1.303333, 2.8555, 2.37, 701, 463, 3.989, 3.85, 14400.5, 15.79, 11.45, 0.915, 7.05, 5.5, 0.023333, 0.0365, 0.0275, 0.123333, 0.4645, 0.12, 15.082, 14.48, 0, 31.8425, 29.1, 7.7325, 3, 5.88, 1.08, 0, 0, 0, 32];
float[] testInput = new float[input.length];
for(int i = 0, total = input.length; i < total; i++){
testInput[i] = new Double(input[i]).floatValue();
}
Booster booster = XGBoost.loadModel("${model}");
//One row, 41 columns
DMatrix testMat = new DMatrix(testInput, 1, 41, Float.NaN);
float[][] predicts = booster.predict(testMat);
3. Instability introduced by missing values in XGBoost on Spark source
However, things aren't that simple.
There is also hidden missing value processing logic in Spark ML: SparseVector, which is a sparse vector.
Both SparseVector and DenseVector are used to represent a vector, and the difference between them is simply the storage structure.
DenseVector is the normal Vector store, where each value in the Vector is stored sequentially.
SparseVector, on the other hand, is a sparse representation for storing data in scenes where the values of 0 are very large in vectors.
The SparseVector is stored by simply recording all non-zero values, ignoring all zero values.Specifically, one array records the locations of all non-zero values, and the other records the values corresponding to those locations.With these two arrays, plus the total length of the current vector, you can restore the original array.
Therefore, SparseVector can save a lot of storage space for a very large set of data with 0 values.
A sample SparseVector storage is shown in the following figure:
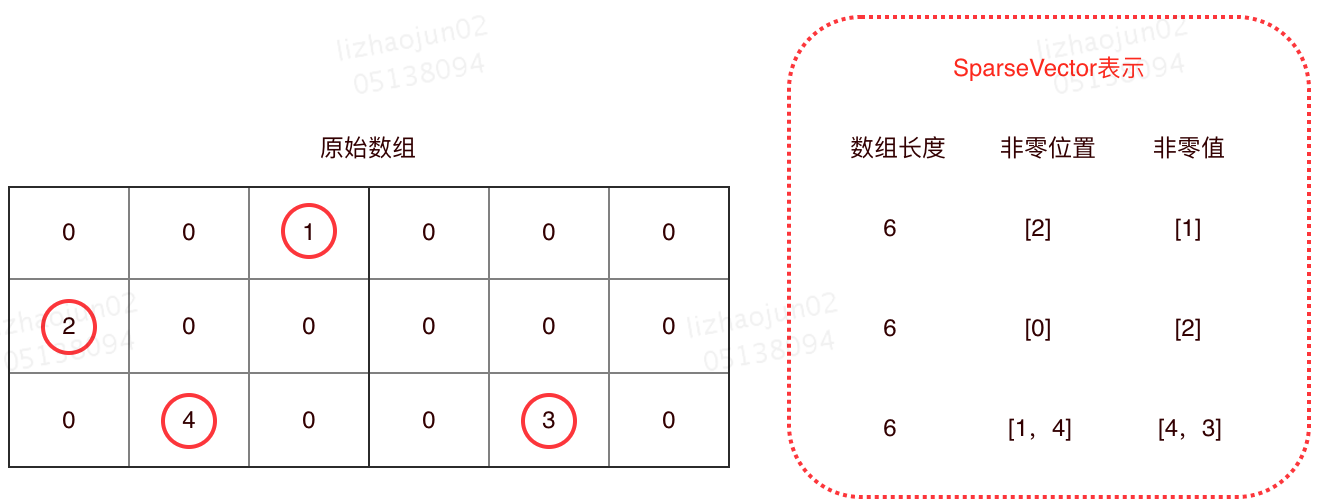
As shown in the figure above, the SparseVector does not save the part of the array that has a value of 0, only records non-zero values.Therefore, a location with a value of 0 does not actually take up storage space.The code below is the implementation code for VectorAssembler in Spark ML, and as you can see from the code, if the value is 0, it is not recorded in SpaseVector.
private[feature] def assemble(vv: Any*): Vector = {
val indices = ArrayBuilder.make[Int]
val values = ArrayBuilder.make[Double]
var cur = 0
vv.foreach {
case v: Double =>
//0 Do not save
if (v != 0.0) {
indices += cur
values += v
}
cur += 1
case vec: Vector =>
vec.foreachActive { case (i, v) =>
//0 Do not save
if (v != 0.0) {
indices += cur + i
values += v
}
}
cur += vec.size
case null =>
throw new SparkException("Values to assemble cannot be null.")
case o =>
throw new SparkException(s"$o of type ${o.getClass.getName} is not supported.")
}
Vectors.sparse(cur, indices.result(), values.result()).compressed
}
A value that does not occupy storage space is also a missing value in a sense.SparseVector is used by all algorithm components, including XGBoost on Spark, as a format for saving arrays in Spark ML.In fact, XGBoost on Spark does treat 0 values in Sparse Vector as missing values:
val instances: RDD[XGBLabeledPoint] = dataset.select(
col($(featuresCol)),
col($(labelCol)).cast(FloatType),
baseMargin.cast(FloatType),
weight.cast(FloatType)
).rdd.map { case Row(features: Vector, label: Float, baseMargin: Float, weight: Float) =>
val (indices, values) = features match {
//SparseVector format, just put non-zero values into the XGBoost calculation
case v: SparseVector => (v.indices, v.values.map(_.toFloat))
case v: DenseVector => (null, v.values.map(_.toFloat))
}
XGBLabeledPoint(label, indices, values, baseMargin = baseMargin, weight = weight)
}
What can XGBoost on Spark cause instability when it considers 0 in SparseVector as a missing value?
Essentially, the storage of the Vector type in Spark ML is optimized, and it automatically chooses whether to store as a SparseVector or a DenseVector based on what is in the Vector array.That is, a field of type Vector has two save formats for the same column when it is saved in SparseVector and DenseVector.And for a column in a data, both formats exist at the same time, with some rows represented by Sparse and some rows represented by Dense.Choose which format to use to represent the calculation from the following code:
/**
* Returns a vector in either dense or sparse format, whichever uses less storage.
*/
@Since("2.0.0")
def compressed: Vector = {
val nnz = numNonzeros
// A dense vector needs 8 * size + 8 bytes, while a sparse vector needs 12 * nnz + 20 bytes.
if (1.5 * (nnz + 1.0) < size) {
toSparse
} else {
toDense
}
}
In the XGBoost on Spark scenario, Float.NaN is the default missing value.If a row in a dataset is stored as a DenseVector, the missing value for that row is Float.NaN when it is actually executed.If a row storage structure in a dataset is a SparseVector, the missing values for that row are Float.NaN and 0 because XGBoost on Spark only uses non-zero values in the SparseVector.
That is, if a row of data in the dataset is suitable for storage as a DenseVector, the missing value for that row is Float.NaN when XGBoost is processed.If the row data is suitable for storage as a SparseVector, the missing values for the row are Float.NaN and 0 when XGBoost processes it.
That is, some of the data in the dataset will have Float.NaN and 0 as the missing values, while others will have Float.NaN as the missing values!That is, in XGBoost on Spark, 0 values have two meanings because of the underlying data storage structure, which is entirely determined by the dataset.
Because only one missing value can be set for online Serving, a test set selected in the SparseVector format may result in results that do not match those expected for online Serving.
4. Problem solving
Checked the latest source code for XGBoost on Spark and still did not solve the problem.
Get back to XGBoost on Spark and modify our own XGBoost on Spark code.
val instances: RDD[XGBLabeledPoint] = dataset.select(
col($(featuresCol)),
col($(labelCol)).cast(FloatType),
baseMargin.cast(FloatType),
weight.cast(FloatType)
).rdd.map { case Row(features: Vector, label: Float, baseMargin: Float, weight: Float) =>
//The return format of the original code needs to be modified here
val values = features match {
//SparseVector data, converted to Dense first
case v: SparseVector => v.toArray.map(_.toFloat)
case v: DenseVector => v.values.map(_.toFloat)
}
XGBLabeledPoint(label, null, values, baseMargin = baseMargin, weight = weight)
}
/**
* Converts a [[Vector]] to a data point with a dummy label.
*
* This is needed for constructing a [[ml.dmlc.xgboost4j.scala.DMatrix]]
* for prediction.
*/
def asXGB: XGBLabeledPoint = v match {
case v: DenseVector =>
XGBLabeledPoint(0.0f, null, v.values.map(_.toFloat))
case v: SparseVector =>
//SparseVector data, converted to Dense first
XGBLabeledPoint(0.0f, null, v.toArray.map(_.toFloat))
}
It is an unexpected pleasure that the problem has been solved and that the evaluation index of the model trained with the new code will be slightly improved.
I hope this article will be helpful to the students who have encountered the missing values of XGBoost, and welcome you to exchange and discuss.
Introduction to the Author
- Zhaojun, technical expert of algorithm platform team of Meituan Distribution Department.
Recruitment Information
The algorithm platform team of Meituan Distribution Department is responsible for the construction of Turing platform, a one-stop large-scale machine learning platform for Meituan.Around the entire lifecycle of the algorithm, a visual drag-and-drop method is used to define the model training and prediction process, provide powerful model management, online model prediction and task conquest capabilities, and provide support for multidimensional stereoscopic AB shunting and online effect evaluation.The mission of the team is to provide a unified, end-to-end, one-stop self-service platform for algorithmic students, to help them reduce the complexity of algorithmic development and improve the efficiency of algorithmic iteration.