preface
Hello! buddy!
Thank you very much for reading Haihong's article. If there are mistakes in the article, you are welcome to point out ~
Self introduction ˊ ᵕ ˋ) ੭
Nickname: Hai Hong
Label: program ape | C + + player | student
Introduction: I got acquainted with programming in C language and then transferred to computer major. I was lucky to have won some national and provincial awards... And I have been guaranteed to study. Currently learning C++/Linux/Python
Learning experience: solid foundation + taking more notes + typing more codes + thinking more + learning English well!
Beginner Python Xiaobai stage
The article is only used as your own learning notes for knowledge system establishment and review
The problem is not to learn one more problem and understand one more problem
Know it, know why!
Previous articles
Python mathematical modeling series (1): linear programming of programming problems
Python mathematical modeling series (2): integer programming of programming problems
Python mathematical modeling series (3): nonlinear programming of planning problems
Python mathematical modeling series (4): numerical approximation
1. Classification of differential equations
Differential equation is an equation used to describe the relationship between a certain kind of function and its derivative, and its solution is a function conforming to the equation.
Differential equations can be divided into ordinary differential equations and partial differential equations according to the number of independent variables
ordinary differential equation (ODE)
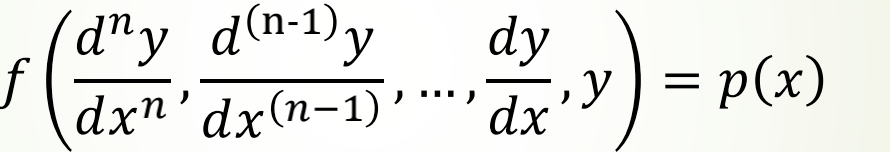
Partial differential equation (more than two independent variables)
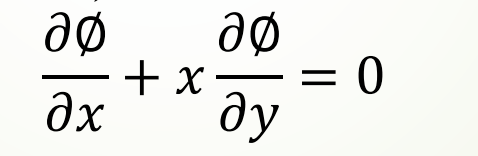
2. Analytical solutions of differential equations
ODE (ordinary differential equation) with analytical solution can be solved by using SymPy library
Taking the second-order ODE of a damped harmonic oscillator as an example, the expression is:
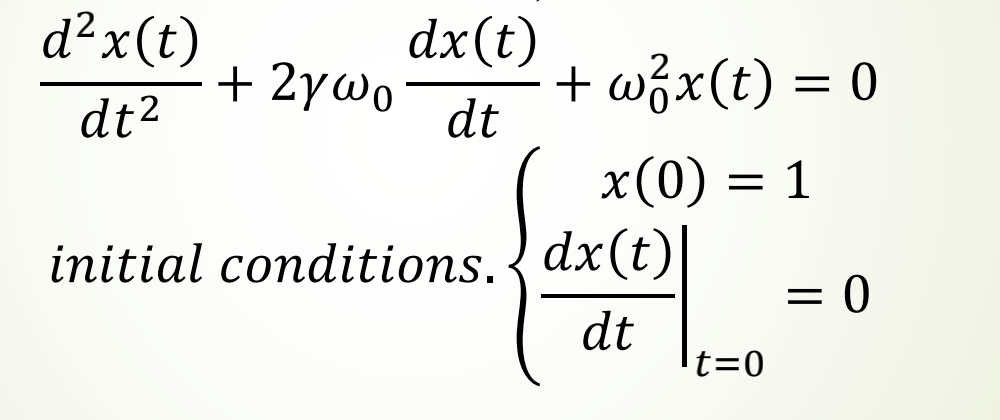
Demo code
import sympy
def apply_ics(sol, ics, x, known_params):
free_params = sol.free_symbols - set(known_params)
eqs = [(sol.lhs.diff(x, n) - sol.rhs.diff(x, n)).subs(x, 0).subs(ics) for n in range(len(ics))]
sol_params = sympy.solve(eqs, free_params)
return sol.subs(sol_params)
# Initialize print environment
sympy.init_printing()
# Mark the parameters and they are all positive
t, omega0, gamma = sympy.symbols("t, omega_0, gamma", positive=True)
# Mark x is a differential function, not a variable
x = sympy.Function("x")
# The general solution is obtained by diff() and dsolve
# ode differential equation is the part to the left of the equal sign and 0 to the right of the equal sign
ode = x(t).diff(t, 2) + 2 * gamma * omega0 * x(t).diff(t) + omega0 ** 2 * x(t)
ode_sol = sympy.dsolve(ode)
# Initial condition: dictionary matching
ics = {x(0): 1, x(t).diff(t).subs(t, 0): 0}
x_t_sol = apply_ics(ode_sol, ics, t, [omega0, gamma])
sympy.pprint(x_t_sol)
Operation results:
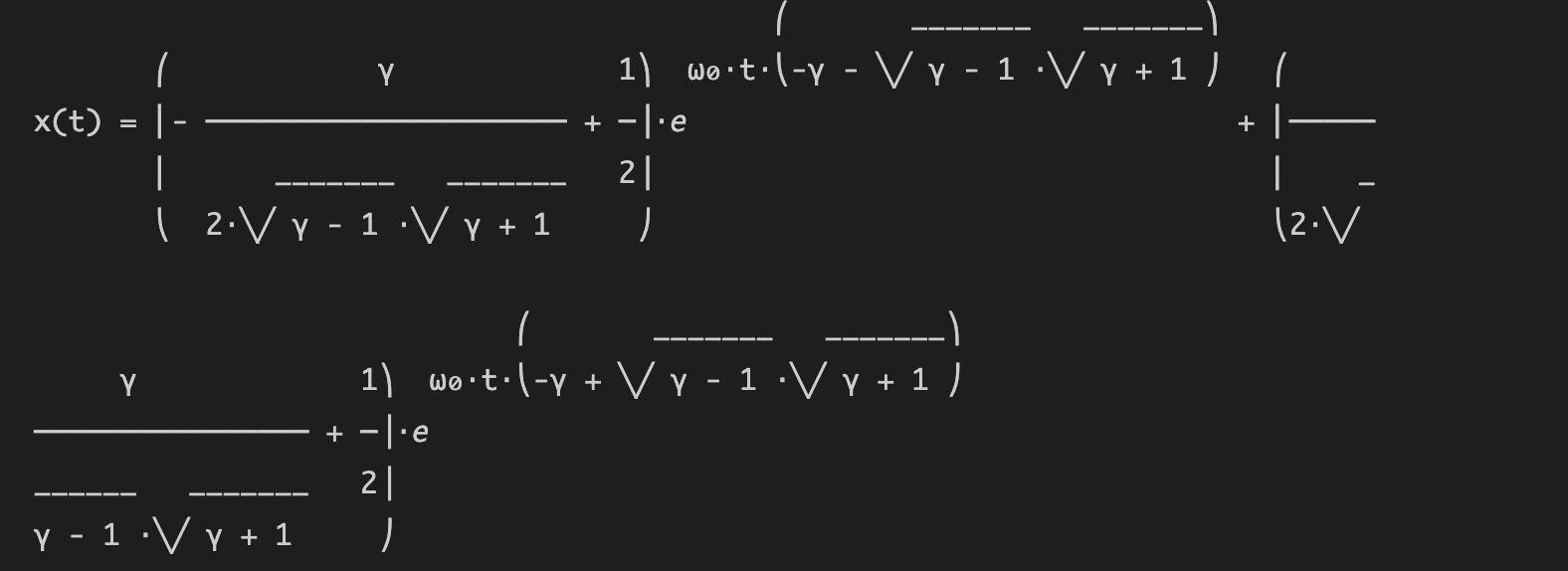
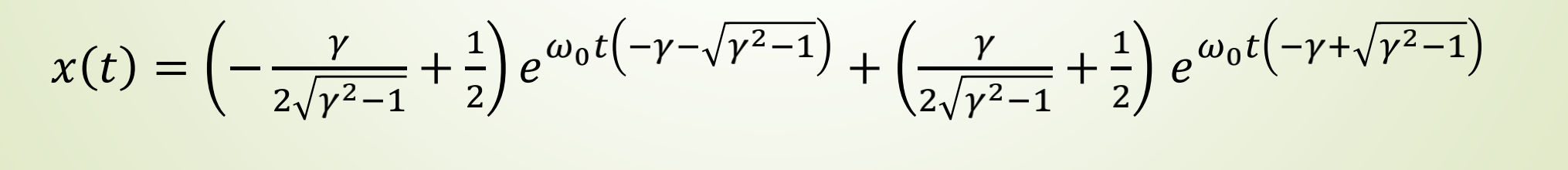
3. Numerical solution of differential equation
When ODE cannot obtain an analytical solution, you can use integrate. In scipy Odeint seeks the numerical solution to explore some properties of its solution, supplemented by visualization, which can intuitively show the functional expression of ODE solution.
Take the following first-order nonlinear (because the power of function y is 2) ODE as an example:
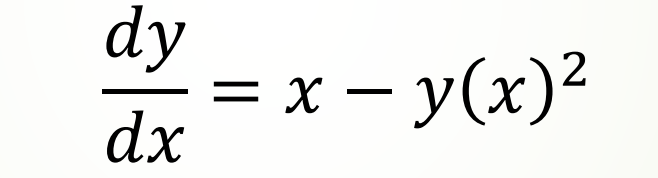
Now we use odeint to find its numerical solution
3.1 field line diagram and numerical solution
Demo code
import numpy as np
from scipy import integrate
import matplotlib.pyplot as plt
import sympy
def plot_direction_field(x, y_x, f_xy, x_lim=(-5, 5), y_lim=(-5, 5), ax=None):
f_np = sympy.lambdify((x, y_x), f_xy, 'numpy')
x_vec = np.linspace(x_lim[0], x_lim[1], 20)
y_vec = np.linspace(y_lim[0], y_lim[1], 20)
if ax is None:
_, ax = plt.subplots(figsize=(4, 4))
dx = x_vec[1] - x_vec[0]
dy = y_vec[1] - y_vec[0]
for m, xx in enumerate(x_vec):
for n, yy in enumerate(y_vec):
Dy = f_np(xx, yy) * dx
Dx = 0.8 * dx**2 / np.sqrt(dx**2 + Dy**2)
Dy = 0.8 * Dy*dy / np.sqrt(dx**2 + Dy**2)
ax.plot([xx - Dx/2, xx + Dx/2], [yy - Dy/2, yy + Dy/2], 'b', lw=0.5)
ax.axis('tight')
ax.set_title(r"$%s$" %(sympy.latex(sympy.Eq(y_x.diff(x), f_xy))), fontsize=18)
return ax
x = sympy.symbols('x')
y = sympy.Function('y')
f = x-y(x)**2
f_np = sympy.lambdify((y(x), x), f)
## put variables (y(x), x) into lambda function f.
y0 = 1
xp = np.linspace(0, 5, 100)
yp = integrate.odeint(f_np, y0, xp)
## solve f_np with initial conditons y0, and x ranges as xp.
xn = np.linspace(0, -5, 100)
yn = integrate.odeint(f_np, y0, xn)
fig, ax = plt.subplots(1, 1, figsize=(4, 4))
plot_direction_field(x, y(x), f, ax=ax)
## plot direction field of function f
ax.plot(xn, yn, 'b', lw=2)
ax.plot(xp, yp, 'r', lw=2)
plt.show()
Operation results:
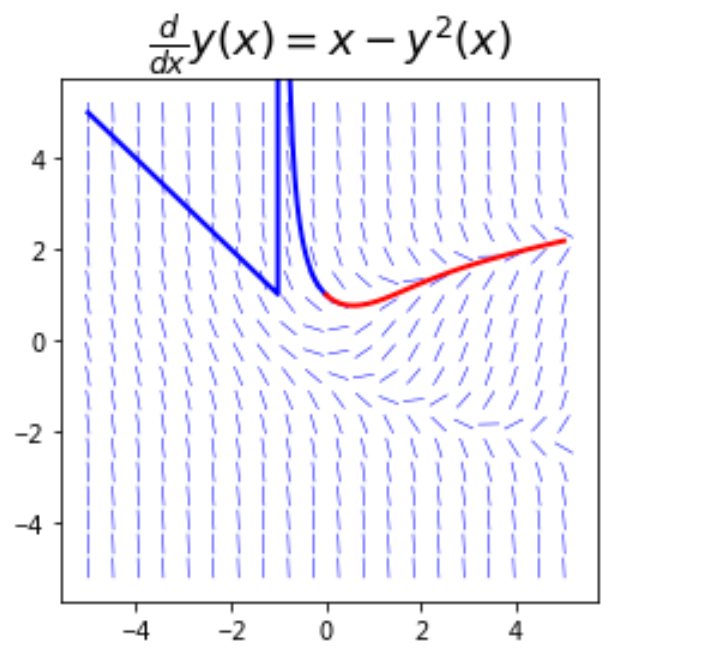
3.2 Lorentz curve and numerical solution
Taking solving the Lorentz curve as an example, the following equations represent the speed of the curve in xyz three directions. Given an initial point, the corresponding Lorentz curve can be drawn:
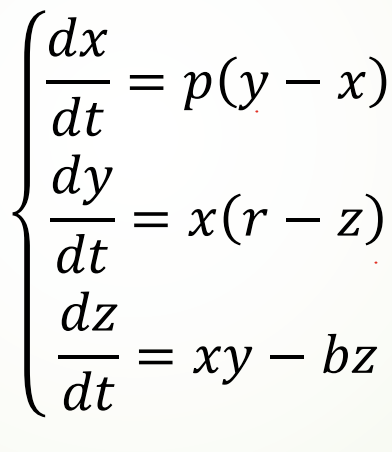
Demo code
import numpy as np
from scipy.integrate import odeint
from mpl_toolkits.mplot3d import Axes3D
import matplotlib.pyplot as plt
def dmove(Point, t, sets):
p, r, b = sets
x, y, z = Point
return np.array([p * (y - x), x * (r - z), x * y - b * z])
t = np.arange(0, 30, 0.001)
P1 = odeint(dmove, (0., 1., 0.), t, args=([10., 28., 3.],))
P2 = odeint(dmove, (0., 1.01, 0.), t, args=([10., 28., 3.],))
fig = plt.figure()
ax = Axes3D(fig)
ax.plot(P1[:, 0], P1[:, 1], P1[:, 2])
ax.plot(P2[:, 0], P2[:, 1], P2[:, 2])
plt.show()
Operation results:
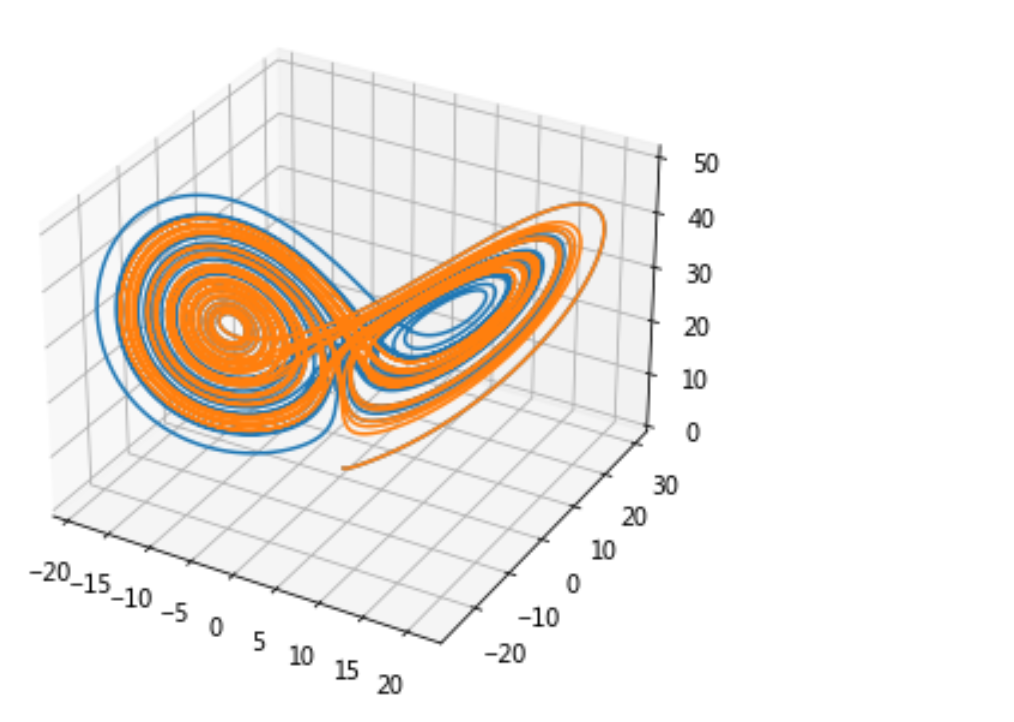
4. Infectious disease model

Model 1: Si model
import scipy.integrate as spi
import numpy as np
import matplotlib.pyplot as plt
# N is the total number of people
N = 10000
# β Is the infection rate coefficient
beta = 0.25
# gamma is the recovery coefficient
gamma = 0
# I_0 is the initial number of infected persons
I_0 = 1
# S_0 is the initial number of susceptible people
S_0 = N - I_0
# T is the propagation time
T = 150
# INI is the array in the initial state
INI = (S_0,I_0)
def funcSI(inivalue,_):
Y = np.zeros(2)
X = inivalue
# Susceptible individual change
Y[0] = - (beta * X[0] * X[1]) / N + gamma * X[1]
# Individual change of infection
Y[1] = (beta * X[0] * X[1]) / N - gamma * X[1]
return Y
T_range = np.arange(0,T + 1)
RES = spi.odeint(funcSI,INI,T_range)
plt.plot(RES[:,0],color = 'darkblue',label = 'Susceptible',marker = '.')
plt.plot(RES[:,1],color = 'red',label = 'Infection',marker = '.')
plt.title('SI Model')
plt.legend()
plt.xlabel('Day')
plt.ylabel('Number')
plt.show()
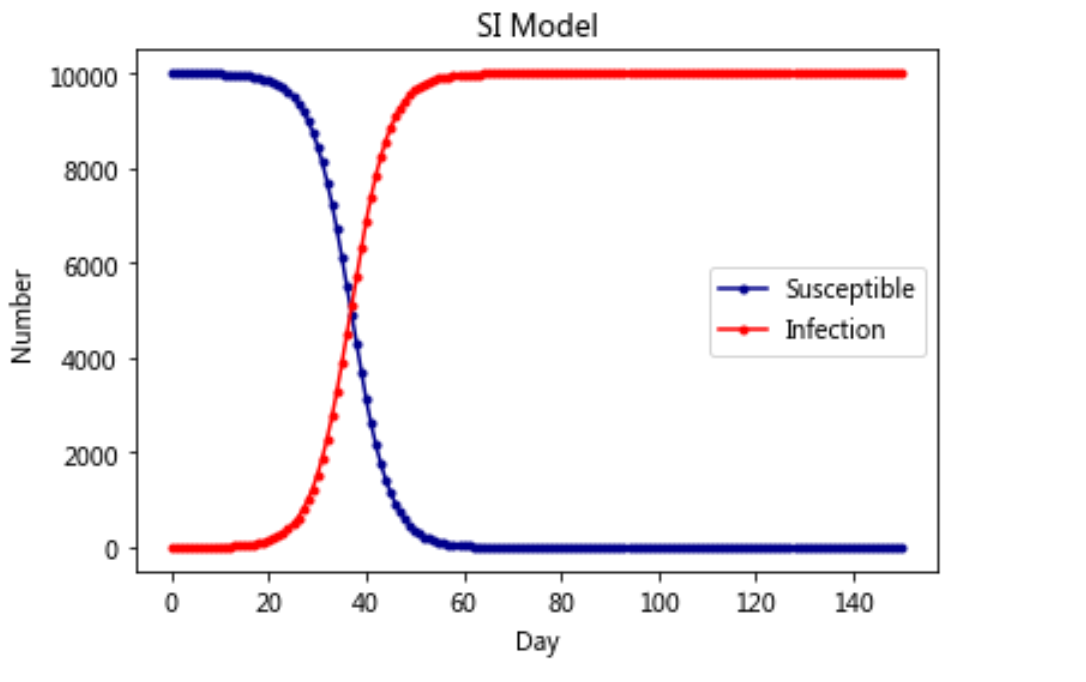
Model 2: SIS model
import scipy.integrate as spi
import numpy as np
import matplotlib.pyplot as plt
# N is the total number of people
N = 10000
# β Is the infection rate coefficient
beta = 0.25
# gamma is the recovery coefficient
gamma = 0.05
# I_0 is the initial number of infected persons
I_0 = 1
# S_0 is the initial number of susceptible people
S_0 = N - I_0
# T is the propagation time
T = 150
# INI is the array in the initial state
INI = (S_0,I_0)
def funcSIS(inivalue,_):
Y = np.zeros(2)
X = inivalue
# Susceptible individual change
Y[0] = - (beta * X[0]) / N * X[1] + gamma * X[1]
# Individual change of infection
Y[1] = (beta * X[0] * X[1]) / N - gamma * X[1]
return Y
T_range = np.arange(0,T + 1)
RES = spi.odeint(funcSIS,INI,T_range)
plt.plot(RES[:,0],color = 'darkblue',label = 'Susceptible',marker = '.')
plt.plot(RES[:,1],color = 'red',label = 'Infection',marker = '.')
plt.title('SIS Model')
plt.legend()
plt.xlabel('Day')
plt.ylabel('Number')
plt.show()
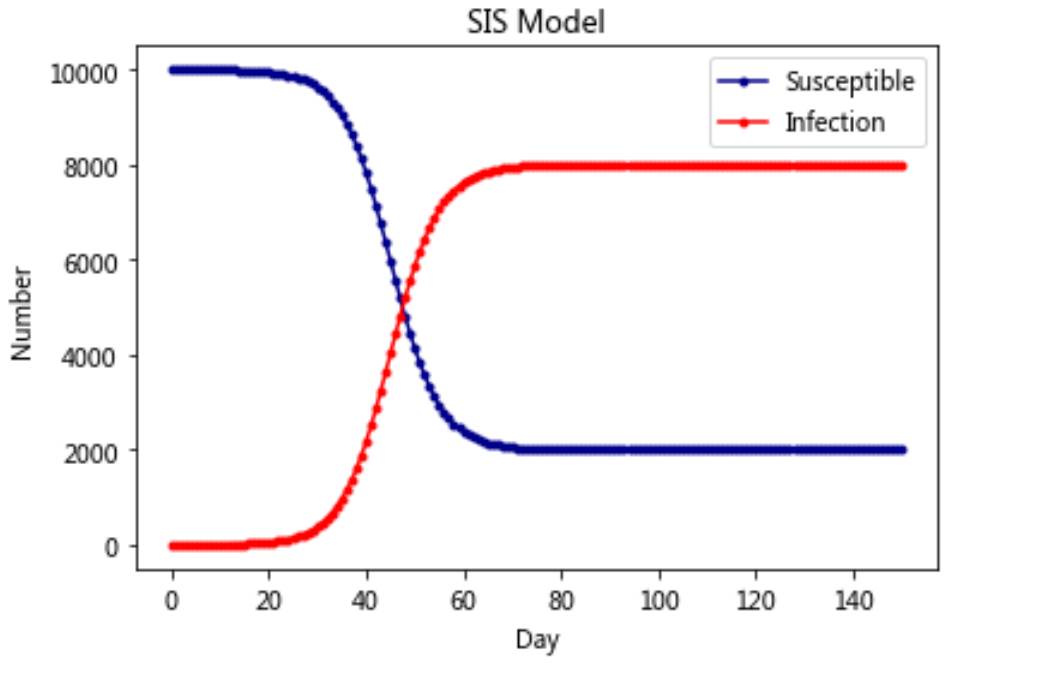
Model 3: SIR model
import scipy.integrate as spi
import numpy as np
import matplotlib.pyplot as plt
# N is the total number of people
N = 10000
# β Is the infection rate coefficient
beta = 0.25
# gamma is the recovery coefficient
gamma = 0.05
# I_0 is the initial number of infected persons
I_0 = 1
# R_0 is the initial number of healers
R_0 = 0
# S_0 is the initial number of susceptible people
S_0 = N - I_0 - R_0
# T is the propagation time
T = 150
# INI is the array in the initial state
INI = (S_0,I_0,R_0)
def funcSIR(inivalue,_):
Y = np.zeros(3)
X = inivalue
# Susceptible individual change
Y[0] = - (beta * X[0] * X[1]) / N
# Individual change of infection
Y[1] = (beta * X[0] * X[1]) / N - gamma * X[1]
# Cure individual changes
Y[2] = gamma * X[1]
return Y
T_range = np.arange(0,T + 1)
RES = spi.odeint(funcSIR,INI,T_range)
plt.plot(RES[:,0],color = 'darkblue',label = 'Susceptible',marker = '.')
plt.plot(RES[:,1],color = 'red',label = 'Infection',marker = '.')
plt.plot(RES[:,2],color = 'green',label = 'Recovery',marker = '.')
plt.title('SIR Model')
plt.legend()
plt.xlabel('Day')
plt.ylabel('Number')
plt.show()
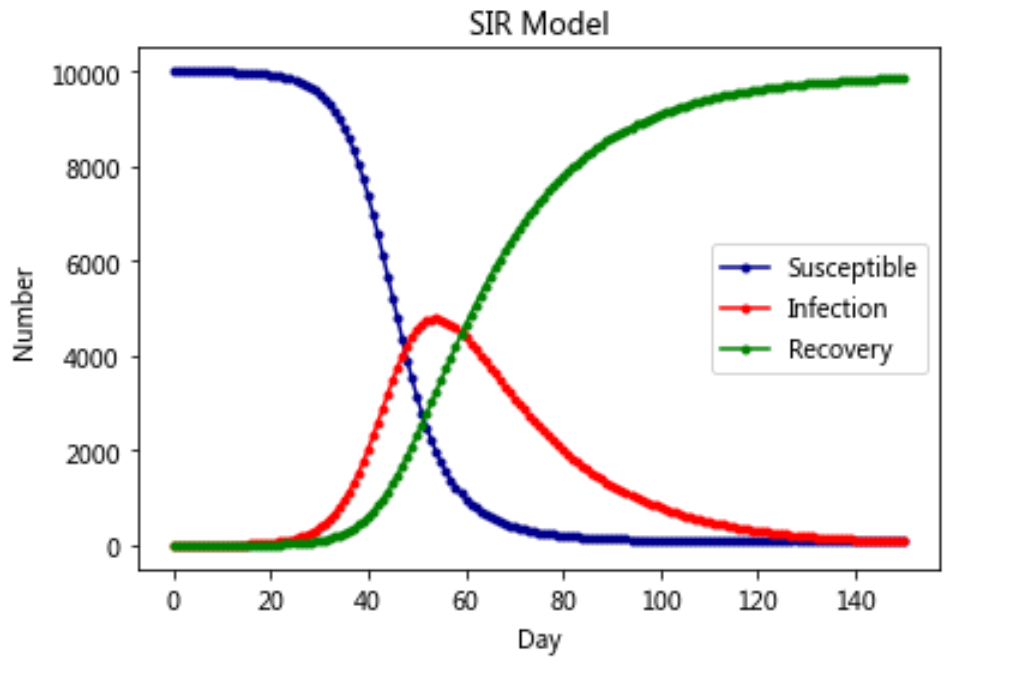
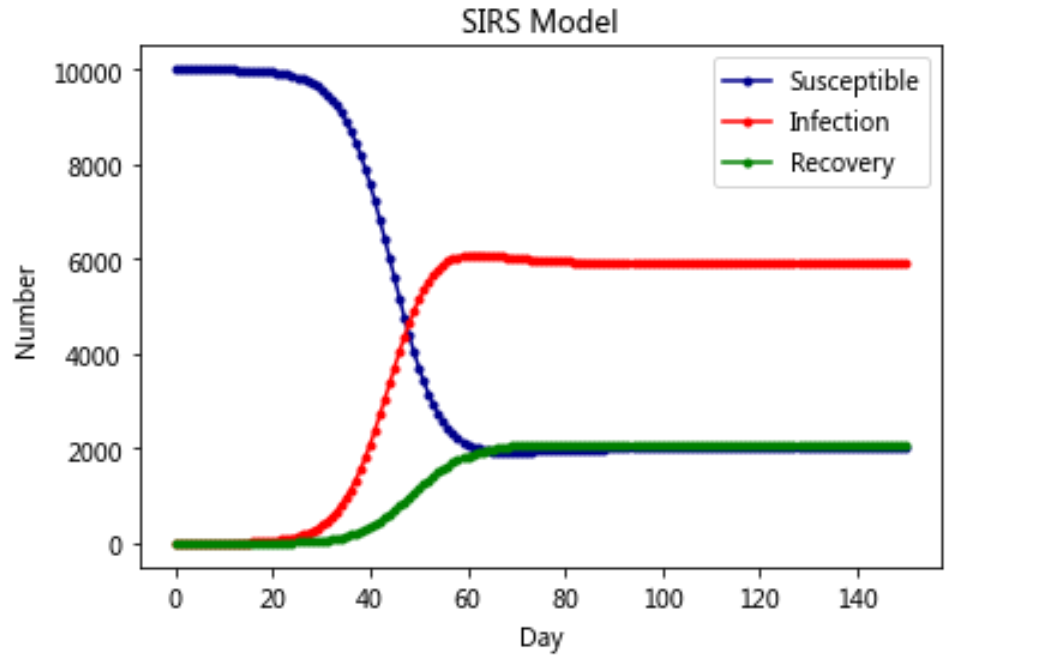
Model 4: SIRS model
import scipy.integrate as spi
import numpy as np
import matplotlib.pyplot as plt
# N is the total number of people
N = 10000
# β Is the infection rate coefficient
beta = 0.25
# gamma is the recovery coefficient
gamma = 0.05
# Ts is the antibody duration
Ts = 7
# I_0 is the initial number of infected persons
I_0 = 1
# R_0 is the initial number of healers
R_0 = 0
# S_0 is the initial number of susceptible people
S_0 = N - I_0 - R_0
# T is the propagation time
T = 150
# INI is the array in the initial state
INI = (S_0,I_0,R_0)
def funcSIRS(inivalue,_):
Y = np.zeros(3)
X = inivalue
# Susceptible individual change
Y[0] = - (beta * X[0] * X[1]) / N + X[2] / Ts
# Individual change of infection
Y[1] = (beta * X[0] * X[1]) / N - gamma * X[1]
# Cure individual changes
Y[2] = gamma * X[1] - X[2] / Ts
return Y
T_range = np.arange(0,T + 1)
RES = spi.odeint(funcSIRS,INI,T_range)
plt.plot(RES[:,0],color = 'darkblue',label = 'Susceptible',marker = '.')
plt.plot(RES[:,1],color = 'red',label = 'Infection',marker = '.')
plt.plot(RES[:,2],color = 'green',label = 'Recovery',marker = '.')
plt.title('SIRS Model')
plt.legend()
plt.xlabel('Day')
plt.ylabel('Number')
plt.show()
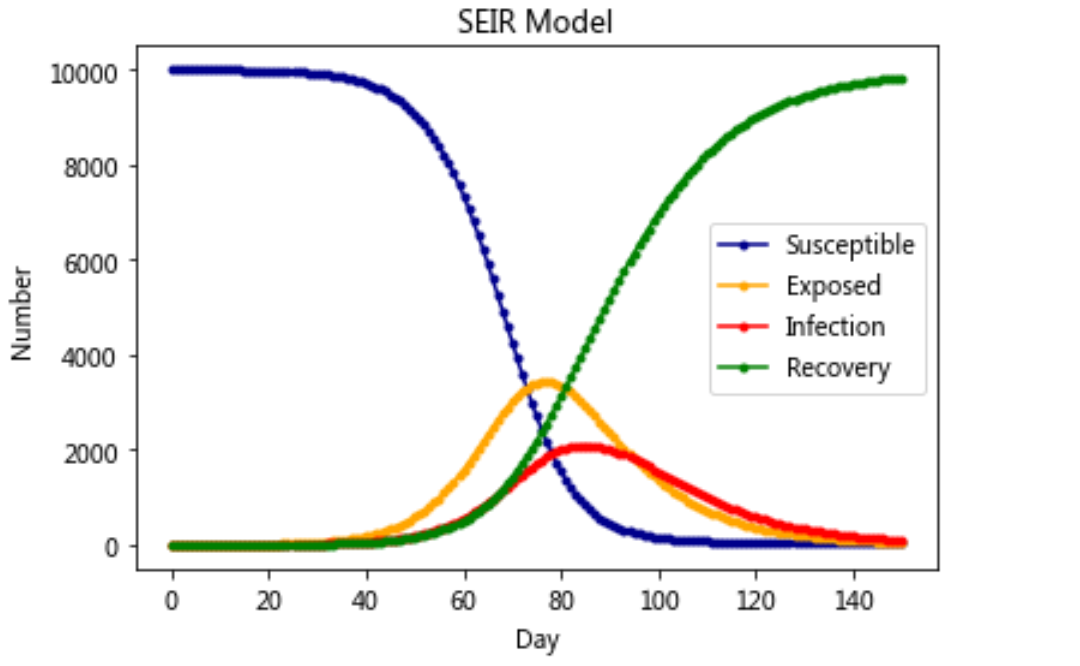
Model 5: SEIR model
import scipy.integrate as spi
import numpy as np
import matplotlib.pyplot as plt
# N is the total number of people
N = 10000
# β Is the infection rate coefficient
beta = 0.6
# gamma is the recovery coefficient
gamma = 0.1
# Te is the disease incubation period
Te = 14
# I_0 is the initial number of infected persons
I_0 = 1
# E_0 is the initial number of lurks
E_0 = 0
# R_0 is the initial number of healers
R_0 = 0
# S_0 is the initial number of susceptible people
S_0 = N - I_0 - E_0 - R_0
# T is the propagation time
T = 150
# INI is the array in the initial state
INI = (S_0,E_0,I_0,R_0)
def funcSEIR(inivalue,_):
Y = np.zeros(4)
X = inivalue
# Susceptible individual change
Y[0] = - (beta * X[0] * X[2]) / N
# Latent individual change
Y[1] = (beta * X[0] * X[2]) / N - X[1] / Te
# Individual change of infection
Y[2] = X[1] / Te - gamma * X[2]
# Cure individual changes
Y[3] = gamma * X[2]
return Y
T_range = np.arange(0,T + 1)
RES = spi.odeint(funcSEIR,INI,T_range)
plt.plot(RES[:,0],color = 'darkblue',label = 'Susceptible',marker = '.')
plt.plot(RES[:,1],color = 'orange',label = 'Exposed',marker = '.')
plt.plot(RES[:,2],color = 'red',label = 'Infection',marker = '.')
plt.plot(RES[:,3],color = 'green',label = 'Recovery',marker = '.')
plt.title('SEIR Model')
plt.legend()
plt.xlabel('Day')
plt.ylabel('Number')
plt.show()
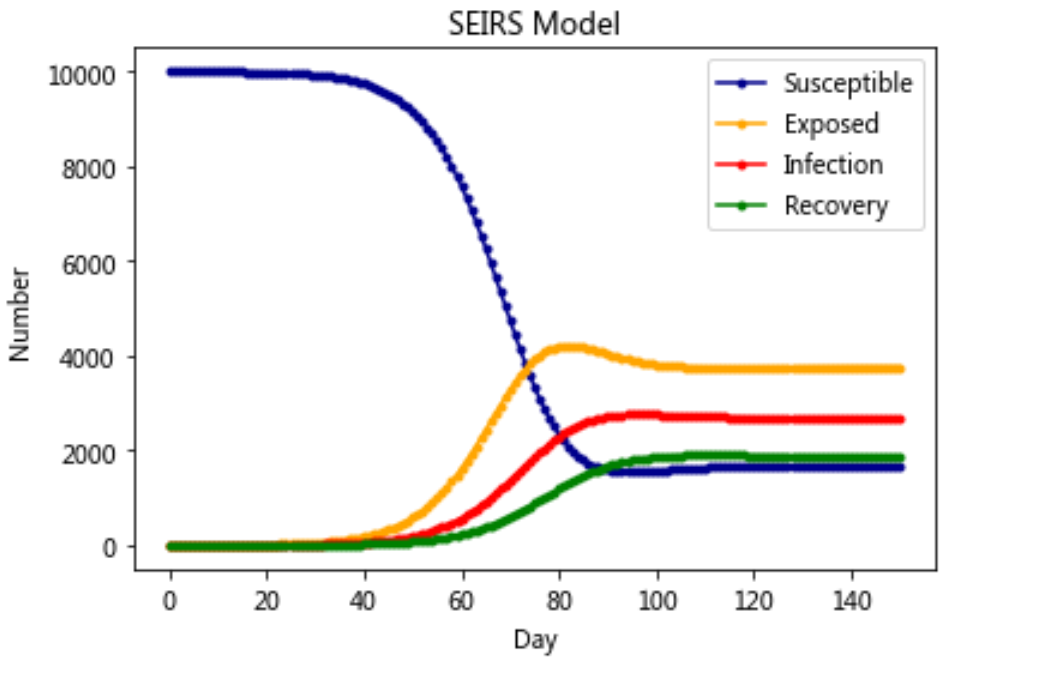
Model 6: SEIRS model
import scipy.integrate as spi
import numpy as np
import matplotlib.pyplot as plt
# N is the total number of people
N = 10000
# β Is the infection rate coefficient
beta = 0.6
# gamma is the recovery coefficient
gamma = 0.1
# Ts is the antibody duration
Ts = 7
# Te is the disease incubation period
Te = 14
# I_0 is the initial number of infected persons
I_0 = 1
# E_0 is the initial number of lurks
E_0 = 0
# R_0 is the initial number of healers
R_0 = 0
# S_0 is the initial number of susceptible people
S_0 = N - I_0 - E_0 - R_0
# T is the propagation time
T = 150
# INI is the array in the initial state
INI = (S_0,E_0,I_0,R_0)
def funcSEIRS(inivalue,_):
Y = np.zeros(4)
X = inivalue
# Susceptible individual change
Y[0] = - (beta * X[0] * X[2]) / N + X[3] / Ts
# Latent individual change
Y[1] = (beta * X[0] * X[2]) / N - X[1] / Te
# Individual change of infection
Y[2] = X[1] / Te - gamma * X[2]
# Cure individual changes
Y[3] = gamma * X[2] - X[3] / Ts
return Y
T_range = np.arange(0,T + 1)
RES = spi.odeint(funcSEIRS,INI,T_range)
plt.plot(RES[:,0],color = 'darkblue',label = 'Susceptible',marker = '.')
plt.plot(RES[:,1],color = 'orange',label = 'Exposed',marker = '.')
plt.plot(RES[:,2],color = 'red',label = 'Infection',marker = '.')
plt.plot(RES[:,3],color = 'green',label = 'Recovery',marker = '.')
plt.title('SEIRS Model')
plt.legend()
plt.xlabel('Day')
plt.ylabel('Number')
plt.show()
epilogue
reference resources:
- https://www.bilibili.com/video/BV12h411d7Dm
- https://zhuanlan.zhihu.com/p/104091330
Learning source: station B and its classroom PPT, in which the code is reproduced
The article is only used as a learning note to record a process from 0 to 1
I hope it will help you. If you have any mistakes, you are welcome to correct them ~
I'm Hai Hongyu ˊ ᵕ ˋ) ੭
If you think it's OK, please like it
Thank you for your support ❤️
