In Tune, some hyperparametric optimization algorithms are written as "scheduling algorithms". These trial schedulers can terminate the adverse test, suspend the test, clone the test and change the super parameters of the running test in advance.
All trial schedulers accept a metric, which is the value returned in your trainable results dictionary, according to the maximize or minimize mode.
tune.run( ... , scheduler=Scheduler(metric="accuracy", mode="max"))
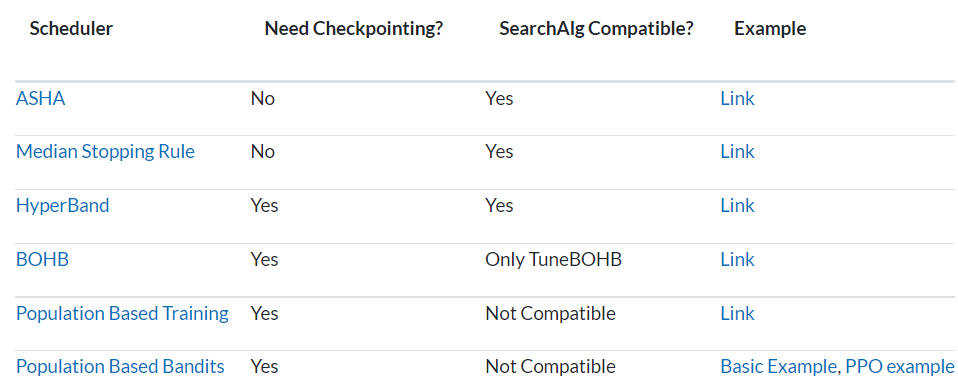
When using schedulers, you may encounter compatibility problems. Some schedulers cannot be used with search algorithms, and the implementation of some schedulers needs to rely on checkpoints.
The scheduler can dynamically change the requirements of using resources for adjustment. Resource changing scheduler can be compatible with other schedulers.
Introduction to scheduling algorithms
- Asha scheduler: the easiest scheduler that will actively terminate low-performance experiments.
- Asynchyperband scheduler: realize asynchronous continuous halving, and solve the backward problem faced by the latter based on the Hyperband scheduler.
- Hyperband scheduler: it is recommended to use the ASHA scheduler on this standard hyperband scheduler. Tune's stop criteria will be applied together with hyperband's early stop mechanism.
- MedianStoppingRule: median stop rule. If the performance of one test is lower than the median of other tests at the time point of rice, the test shall be stopped.
- Population based training: population based training. Each trial variant was considered a member of the group and the best performing trials were regularly checked. Low performance trials clone the checkpoints of the best performers and disrupt the configuration to find the best variants together. Different from other super parameter search algorithms, PBT will change the super parameters during training, which can realize very fast super parameter discovery and automatically find a good annealing schedule.
- Populationbasedtraining replay: return visit based on population training. Hyperparametric plan for replay PBT. Strictly speaking, it is not a scheduler. Instead, it is a PBT recovery tool.
- PB2. Based on PBT, the main difference is that PB2 uses Gaussian process model instead of random disturbance to select new hyperparametric configuration** The main motivation is to find promising hyperparameters with a small population size** PB2 trains a group of models in parallel. Periodically, the model with poor performance will clone the model with the best performance, and use GP bandit optimization to re select the super parameters. The GP model is trained to predict the improvement of the next training cycle.
- HyperBandForBOHB: BOHB. Is a variant of hyperband. It is the same as the original implementation of hyperband, but does not implement pipelining or straggler mitigation. It is recommended to be used in combination with the TuneBOHB search algorithm.
Examples of common scheduling algorithms
1 AsyncHyperBandScheduler
import ray
from ray import tune
from ray.tune.schedulers import AsyncHyperBandScheduler
def easy_objective(config):
# Hyperparameters
width, height = config["width"], config["height"]
for step in range(config["steps"]):
# Iterative training function - can be an arbitrary training procedure
intermediate_score = evaluation_fn(step, width, height)
# Feed the score back back to Tune.
tune.report(iterations=step, mean_loss=intermediate_score)
if __name__ == "__main__":
# AsyncHyperBand enables aggressive early stopping of bad trials.
scheduler = AsyncHyperBandScheduler(grace_period=5, max_t=100)
# 'training_iteration' is incremented every time `trainable.step` is called
stopping_criteria = {"training_iteration": 1 if args.smoke_test else 9999}
analysis = tune.run(
easy_objective,
name="asynchyperband_test",
metric="mean_loss",
mode="min",
scheduler=scheduler,
stop=stopping_criteria,
num_samples=20,
verbose=1,
resources_per_trial={
"cpu": 1,
"gpu": 0
},
config={ # Hyperparameter space
"steps": 100,
"width": tune.uniform(10, 100),
"height": tune.uniform(0, 100),
})
print("Best hyperparameters found were: ", analysis.best_config)
2 PB2
import os
import random
import argparse
import pandas as pd
from datetime import datetime
from ray.tune import run, sample_from
from ray.tune.schedulers import PopulationBasedTraining
from ray.tune.schedulers.pb2 import PB2
# Postprocess the perturbed config to ensure it's still valid used if PBT.
def explore(config):
# Ensure we collect enough timesteps to do sgd.
if config["train_batch_size"] < config["sgd_minibatch_size"] * 2:
config["train_batch_size"] = config["sgd_minibatch_size"] * 2
# Ensure we run at least one sgd iter.
if config["lambda"] > 1:
config["lambda"] = 1
config["train_batch_size"] = int(config["train_batch_size"])
return config
if __name__ == "__main__":
parser = argparse.ArgumentParser()
parser.add_argument("--max", type=int, default=1000000)
parser.add_argument("--algo", type=str, default="PPO")
parser.add_argument("--num_workers", type=int, default=4)
parser.add_argument("--num_samples", type=int, default=4)
parser.add_argument("--t_ready", type=int, default=50000)
parser.add_argument("--seed", type=int, default=0)
parser.add_argument(
"--horizon", type=int, default=1600) # make this 1000 for other envs
parser.add_argument("--perturb", type=float, default=0.25) # if using PBT
parser.add_argument("--env_name", type=str, default="BipedalWalker-v2")
parser.add_argument(
"--criteria", type=str,
default="timesteps_total") # "training_iteration", "time_total_s"
parser.add_argument(
"--net", type=str, default="32_32"
) # May be important to use a larger network for bigger tasks.
parser.add_argument("--filename", type=str, default="")
parser.add_argument("--method", type=str, default="pb2") # ['pbt', 'pb2']
parser.add_argument("--save_csv", type=bool, default=False)
args = parser.parse_args()
# bipedalwalker needs 1600
if args.env_name in ["BipedalWalker-v2", "BipedalWalker-v3"]:
horizon = 1600
else:
horizon = 1000
pbt = PopulationBasedTraining(
time_attr=args.criteria,
metric="episode_reward_mean",
mode="max",
perturbation_interval=args.t_ready,
resample_probability=args.perturb,
quantile_fraction=args.perturb, # copy bottom % with top %
# Specifies the search space for these hyperparams
hyperparam_mutations={
"lambda": lambda: random.uniform(0.9, 1.0),
"clip_param": lambda: random.uniform(0.1, 0.5),
"lr": lambda: random.uniform(1e-3, 1e-5),
"train_batch_size": lambda: random.randint(1000, 60000),
},
custom_explore_fn=explore)
pb2 = PB2(
time_attr=args.criteria,
metric="episode_reward_mean",
mode="max",
perturbation_interval=args.t_ready,
quantile_fraction=args.perturb, # copy bottom % with top %
# Specifies the hyperparam search space
hyperparam_bounds={
"lambda": [0.9, 1.0],
"clip_param": [0.1, 0.5],
"lr": [1e-3, 1e-5],
"train_batch_size": [1000, 60000]
})
methods = {"pbt": pbt, "pb2": pb2}
timelog = str(datetime.date(datetime.now())) + "_" + str(
datetime.time(datetime.now()))
args.dir = "{}_{}_{}_Size{}_{}_{}".format(args.algo,
args.filename, args.method,
str(args.num_samples),
args.env_name, args.criteria)
analysis = run(
args.algo,
name="{}_{}_{}_seed{}_{}".format(timelog, args.method, args.env_name,
str(args.seed), args.filename),
scheduler=methods[args.method],
verbose=1,
num_samples=args.num_samples,
stop={args.criteria: args.max},
config={
"env": args.env_name,
"log_level": "INFO",
"seed": args.seed,
"kl_coeff": 1.0,
"num_gpus": 0,
"horizon": horizon,
"observation_filter": "MeanStdFilter",
"model": {
"fcnet_hiddens": [
int(args.net.split("_")[0]),
int(args.net.split("_")[1])
],
"free_log_std": True
},
"num_sgd_iter": 10,
"sgd_minibatch_size": 128,
"lambda": sample_from(lambda spec: random.uniform(0.9, 1.0)),
"clip_param": sample_from(lambda spec: random.uniform(0.1, 0.5)),
"lr": sample_from(lambda spec: random.uniform(1e-3, 1e-5)),
"train_batch_size": sample_from(
lambda spec: random.randint(1000, 60000))
})
all_dfs = analysis.trial_dataframes
names = list(all_dfs.keys())
results = pd.DataFrame()
for i in range(args.num_samples):
df = all_dfs[names[i]]
df = df[[
"timesteps_total", "episodes_total", "episode_reward_mean",
"info/learner/default_policy/cur_kl_coeff"
]]
df["Agent"] = i
results = pd.concat([results, df]).reset_index(drop=True)
if args.save_csv:
if not (os.path.exists("data/" + args.dir)):
os.makedirs("data/" + args.dir)
results.to_csv("data/{}/seed{}.csv".format(args.dir, str(args.seed)))