Application of smac: auto-sklearn
SMAC(Sequential Model-based Algorithm Configuration), an algorithm configuration based on sequence models, is widely used in various automatic machine learning frameworks, such as Microsoft nni. https://github.com/microsoft/nni ), auto-sklearn, etc.The superparametric search module of auto-sklearn is entirely built by Smac algorithm.To understand auto-sklearn, you first need to understand smac.
Here's the smac scenario: AutoML, application body: auto-sklearn
With the recent popularity of artificial intelligence technology, machine learning and in-depth learning have gradually entered people's vision. With the support of various tools (such as Tensorflow, sklearn) open source, machine learning technology has gradually become universal and nationalized.However, taking ordinary tabular data (behavior sample points, listed as features) as an example, it takes a lot of time to try to find the best Pipeline (that is, from data processing, feature processing, to estimator discrimination).Generally speaking, data scientists need to get some basic statistics of the data, make some charts to judge the pattern of the data, select data processing, feature processing methods based on their own experience, and finally choose an estimator (classifier in classification tasks, regressor in regression tasks).However, this process is time consuming and more dependent on the experience of data scientists, raising the threshold for machine learning.
This contradiction has been noticed and the concept of automatic machine learning (AutoML) has been proposed.In 2015, an AutoML framework, auto-sklearn, was launched, drawing attention to: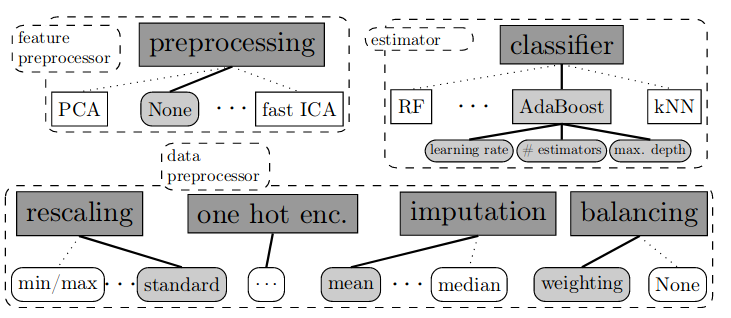
- auto-sklearn Papers and Codes
| paper | http://papers.nips.cc/paper/5872-efficient-and-robust-automated-machine-learning.pdf |
|---|---|
| code | https://github.com/automl/auto-sklearn |
Recently, I was investigating AutoML, so auto-sklearn was investigated.The author found that auto-sklearn is a team of automl ( https://www.automl.org ) Writing, the team used a large number of self-developed libraries to build this automated machine learning framework. The libraries and libraries are cohesive, isolated, and the call relationship is a directed acyclic graph: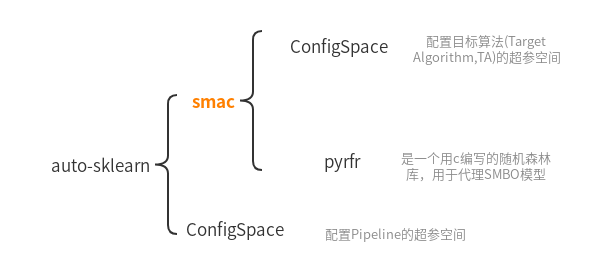
The orange font-marked smac is our hero today.After analyzing the project architecture of auto-sklearn, I downloaded the source code for smac ( https://github.com/automl/SMAC3 ), and begin to analyze it.
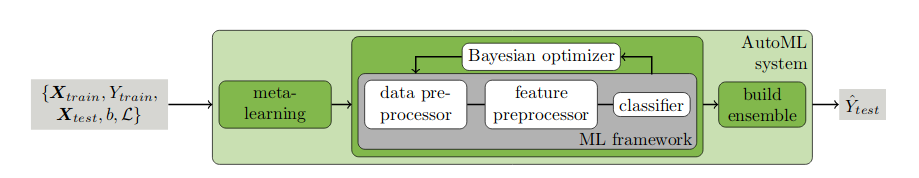 In the AutoML system, the most critical is to give a superparameter that is expected to be optimal under the current conditions based on the current set of hyperparameter combinations (such as the selection of svm kernel function, the selection of penalty factor C) and model performance (such as the accuracy of classification tasks), which is Bayesian Optimizer.Unlike grid searches and random searches, Bayesian optimization finds better hyperparameters in less time.
In the AutoML system, the most critical is to give a superparameter that is expected to be optimal under the current conditions based on the current set of hyperparameter combinations (such as the selection of svm kernel function, the selection of penalty factor C) and model performance (such as the accuracy of classification tasks), which is Bayesian Optimizer.Unlike grid searches and random searches, Bayesian optimization finds better hyperparameters in less time.
SMBO (Sequential Model-Based Optimazation), a model-based sequence optimization, is the most widely used.There are currently three common SMBO algorithms: sequence superparametric optimization based on Gaussian process regression, sequence superparametric optimization based on random forest algorithm proxy (smac), and sequence superparametric optimization based on TPE algorithm proxy.
Smc Quick Start
Start the first example by executing the following command in the terminal
git clone https://github.com/automl/SMAC3 cd SMAC3 && pip install . cd examples python SMAC4HPO_svm.py
Let's see the sample code SMAC4HPO_svm.py.
First, a function svm_from_cfg with input as CFG (superparametric configuration) and output as model representation is defined
def svm_from_cfg(cfg): """ Create a configuration-based SVM Model, and evaluated on Iris dataset by cross-validation Parameters: ----------- cfg: Hyperparameter Configuration (ConfigSpace.ConfigurationSpace.Configuration) Returns: -------- A crossvalidated mean score for the svm on the loaded data-set. svm Average of cross-validation on datasets """ # Filter out key-value pairs with value None by dictionary resolution cfg = {k : cfg[k] for k in cfg if cfg[k]} # We translate boolean values: cfg["shrinking"] = True if cfg["shrinking"] == "true" else False # And for gamma, we set it to a fixed value or to "auto" (if used) if "gamma" in cfg: cfg["gamma"] = cfg["gamma_value"] if cfg["gamma"] == "value" else "auto" cfg.pop("gamma_value", None) # Remove "gamma_value" clf = svm.SVC(**cfg, random_state=42) scores = cross_val_score(clf, iris.data, iris.target, cv=5) return 1-np.mean(scores) # Minimize!
Next, configure the hyperparameter space
# Build Configuration Space which defines all parameters and their ranges cs = ConfigurationSpace() # We define a few possible types of SVM-kernels and add them as "kernel" to our cs kernel = CategoricalHyperparameter("kernel", ["linear", "rbf", "poly", "sigmoid"], default_value="poly") cs.add_hyperparameter(kernel) # There are some hyperparameters shared by all kernels C = UniformFloatHyperparameter("C", 0.001, 1000.0, default_value=1.0) shrinking = CategoricalHyperparameter("shrinking", ["true", "false"], default_value="true") cs.add_hyperparameters([C, shrinking]) # Others are kernel-specific, so we can add conditions to limit the searchspace degree = UniformIntegerHyperparameter("degree", 1, 5, default_value=3) # Only used by kernel poly coef0 = UniformFloatHyperparameter("coef0", 0.0, 10.0, default_value=0.0) # poly, sigmoid cs.add_hyperparameters([degree, coef0]) use_degree = InCondition(child=degree, parent=kernel, values=["poly"]) use_coef0 = InCondition(child=coef0, parent=kernel, values=["poly", "sigmoid"]) cs.add_conditions([use_degree, use_coef0]) # This also works for parameters that are a mix of categorical and values from a range of numbers # For example, gamma can be either "auto" or a fixed float gamma = CategoricalHyperparameter("gamma", ["auto", "value"], default_value="auto") # only rbf, poly, sigmoid gamma_value = UniformFloatHyperparameter("gamma_value", 0.0001, 8, default_value=1) cs.add_hyperparameters([gamma, gamma_value]) # We only activate gamma_value if gamma is set to "value" cs.add_condition(InCondition(child=gamma_value, parent=gamma, values=["value"])) # And again we can restrict the use of gamma in general to the choice of the kernel cs.add_condition(InCondition(child=gamma, parent=kernel, values=["rbf", "poly", "sigmoid"]))
You can see that when configuring the hyperparameter space, in addition to configuring the default value and range of the hyperparameter, you can also configure the Condition s of the hyperparameter, that is, the conditions.For example, svm has four kernel functions ['linear','rbf','poly','sigmoid'], but only ['rbf','poly','sigmoid'] have gamma parameters.
# Scenario object (schema object) scenario = Scenario({"run_obj": "quality", # There are {runtime,quality} options "runcount-limit": 50, # max. number of function evaluations; for this example set to a low number "cs": cs, # configuration space "deterministic": "true" }) # Example call of the function # It returns: Status, Cost, Runtime, Additional Infos def_value = svm_from_cfg(cs.get_default_configuration()) print("Default Value: %.2f" % (def_value)) # Optimize, using a SMAC-object print("Optimizing! Depending on your machine, this might take a few minutes.") smac = SMAC4HPO(scenario=scenario, rng=np.random.RandomState(42), tae_runner=svm_from_cfg) # Incumbent (incumbent, as opposed to challenger) represents the configuration that makes the model perform best incumbent = smac.optimize() # Optimal model performance inc_value = svm_from_cfg(incumbent) print("Optimized Value: %.2f" % (inc_value)) # We can also validate our results (though this makes a lot more sense with instances) smac.validate(config_mode='inc', # We can choose which configurations to evaluate #instance_mode='train+test', # Defines what instances to validate repetitions=100, # Ignored, unless you set "deterministic" to "false" in line 95 n_jobs=1) # How many cores to use in parallel for optimization
We see the last lines of code, first building a Scenario to represent the search configuration, then passing scenario and the target function as parameters into SMAC4HPO to return a smac object and calling optimize() to get an optimal superparameter.
smac project architecture
$ tree smac -L 1
.
├── configspace/
├── epm/
├── facade/
├── initial_design/
├── intensification/
├── optimizer/
├── runhistory/
├── scenario/
├── stats/
├── tae/
└── utils/
| Directory Name | Interpretation |
|---|---|
| configspace | Hyperparameter space |
| epm | Random Forest Agent Model |
| facade | Some decorative categories |
| initial_design | Initialization method of search space |
| intensification | Choosing the optimal configuration in the challenger |
| optimizer | Optimizer, including acquisition, smbo multiple important files |
| runhistory | Recording of run history |
| scenario | Search scenario configuration |
| tae | Target Algorithm Evaluator, Target Algorithm Evaluator |
In the next push, the author will continue to analyze smac source code for you to explore how smac optimizes the hyperparametric space using a random forest agent model.

