Learning objectives:
Convert the data set into Mask, coco annotation and other different formats
1, The coco annotation and Mask of a single instance are converted to each other:
There are many ways of mask conversion. One mask is generated for each instance or one mask is generated for each picture. For beginners, it is better to use the existing tools for the conversion of single instance coco data sets. pycococreator is used for the conversion:
pycococreator project file address:
https://github.com/waspinator/pycococreator
Since the official installation method is on linux, windows can choose to download the project file and open the command prompt in the current project file. If the running environment is in the virtual environment, activate the virtual environment, and then enter the following command:
python setup.py install
For relevant instructions, please refer to the website:
https://patrickwasp.com/create-your-own-coco-style-dataset/
To verify the successful installation, you can enter the following commands:
from pycococreatortools import pycococreatortools
If no error is reported.
1.1 convert each instance into a gray mask
Generate the instance mask according to the marked data.
According to the example of pycococreator, the mask is named as follows:
Here, the naming type of mask generation directly adopts its required naming format, so that the subsequent conversion can be carried out directly.
Relevant codes and notes are shown in the figure:
def objectSingleMask(annFile, classes_names, savePath):
"""
From dimension file annFile Extract compliance category classes_names And save it
Since each image has only one instance, it defaults to grayscale image
:param annFile: json Annotate Document
:param classes_names: The class alias of the mask needs to be extracted
:param savePath: Save path
:return:
"""
# Get COCO_json data
coco = COCO(annFile)
# Get the id of all the required image data - what is the id of the categories I need
classes_ids = coco.getCatIds(catNms=classes_names)
# Take all picture IDs of the union of all categories
# If you want to intersect, you don't need a loop. Directly enter all categories as parameters to get the pictures contained in all categories
imgIds_list = []
# Loop out the pictures corresponding to each category id and obtain the id number of the picture
for idx in classes_ids:
imgidx = coco.getImgIds(catIds=idx) # Put all the picture IDs of this category into a list
imgIds_list += imgidx
print("search id... ", imgidx)
# Remove duplicate images
imgIds_list = list(set(imgIds_list)) # Merge the same picture id corresponding to multiple categories
# Get the information of all images at one time
image_info_list = coco.loadImgs(imgIds_list)
# Generate a mask for each instance of each picture
annotation_id = 1
for imageinfo in image_info_list:
for singleClassIdIndex in range(len(classes_ids)):
singleClassId = classes_ids[singleClassIdIndex]
# Find the label id of the mask
singleClassAnnId = coco.getAnnIds(imgIds=imageinfo['id'], catIds=[singleClassId], iscrowd=None)
# Extract the segmentation data of the mask
singleClassAnnList = coco.loadAnns(singleClassAnnId)
for singleItem in singleClassAnnList:
# Find every instance
singleItemMask = coco.annToMask(singleItem)
# Convert the instance to uint8 of 0 ~ 255 and save it
singleItemMask = (singleItemMask * 255).astype(np.uint8)
# <image_id>_<object_class_name>_<annotation_id>.png
file_name = savePath + '/' + imageinfo['file_name'][:-4] + '_' + \
classes_names[singleClassIdIndex] + \
'_' + '%d' % annotation_id + '.png'
cv2.imwrite(file_name, singleItemMask)
print("Saved mask picture: ", file_name)
annotation_id = annotation_id + 1
Enter directly when the program is running:
# Label file address
jsondir = "data/trainval.json"
# Single mask
singleMaskDir = "mydata/singleMask"
mkr(singleMaskDir)
objectSingleMask(jsondir, ['date', 'fig', 'hazelnut'], singleMaskDir)
Just.
Folder display:

1.2 regenerate the coco annotation file according to the mask of a single instance
# Convert a single mask to a json file
# https://patrickwasp.com/create-your-own-coco-style-dataset/
import datetime
import json
import os
import re
import fnmatch
from PIL import Image
import numpy as np
from pycococreatortools import pycococreatortools
def filter_for_jpeg(root, files):
"""
Extract the picture files in the folder that match the relevant suffix
:param root: File path
:param files: File list under file path
:return: Files that meet the naming criteria
"""
file_types = ['*.jpeg', '*.jpg', '*.png']
file_types = r'|'.join([fnmatch.translate(x) for x in file_types])
files = [os.path.join(root, f) for f in files]
files = [f for f in files if re.match(file_types, f)]
return files
def filter_for_annotations(root, files, image_filename):
"""
Extract the files in the folder that match the relevant suffix coco Note file,
because png The picture is non-destructive, so it is generally used png Format as mask format
:param root: File path
:param files: File list under file path
:param image_filename: coco Picture file corresponding to annotation file
:return:
"""
file_types = ['*.png']
file_types = r'|'.join([fnmatch.translate(x) for x in file_types])
basename_no_extension = os.path.splitext(os.path.basename(image_filename))[0]
file_name_prefix = basename_no_extension + '.*'
files = [os.path.join(root, f) for f in files]
files = [f for f in files if re.match(file_types, f)]
files = [f for f in files if re.match(file_name_prefix, os.path.splitext(os.path.basename(f))[0])]
return files
if __name__ == "__main__":
# Path setting
ROOT_DIR = 'data'
IMAGE_DIR = os.path.join(ROOT_DIR, "images")
ANNOTATION_DIR = "mydata/singleMask"
SAVE_PATH_DIR = "myjson/singleJson.json"
# Relevant information of the note file is optional
INFO = {
"description": "FruitsNuts Dataset",
"url": "https://github.com/fenglingbai/FruitsNutsHandle",
"version": "0.1.0",
"year": 2022,
"contributor": "fenglingbai",
"date_created": datetime.datetime.utcnow().isoformat(' ')
}
LICENSES = [
{
"id": 1,
"name": "None",
"url": "None"
}
]
# Add categories according to your needs
CATEGORIES = [
{
"supercategory": "date",
"id": 1,
"name": "date"
},
{
"supercategory": "fig",
"id": 2,
"name": "fig"
},
{
"supercategory": "hazelnut",
"id": 3,
"name": "hazelnut"
}
]
# Output Dictionary of json file
coco_output = {
"info": INFO,
"licenses": LICENSES,
"categories": CATEGORIES,
"images": [],
"annotations": []
}
# Initialization of annotation label
image_id = 0
segmentation_id = 1
# Original drawing file of filtered data
for root, _, files in os.walk(IMAGE_DIR):
image_files = filter_for_jpeg(root, files)
# Traverse each original drawing file
for image_filename in image_files:
image = Image.open(image_filename)
image_info = pycococreatortools.create_image_info(
image_id, os.path.basename(image_filename), image.size)
coco_output["images"].append(image_info)
# Filter the annotation file corresponding to the original drawing file
for root, _, files in os.walk(ANNOTATION_DIR):
annotation_files = filter_for_annotations(root, files, image_filename)
# Traverse each annotation file
for annotation_filename in annotation_files:
print(annotation_filename)
class_id = [x['id'] for x in CATEGORIES if x['name'] in annotation_filename][0]
category_info = {'id': class_id, 'is_crowd': 'crowd' in image_filename}
binary_mask = np.asarray(Image.open(annotation_filename)
.convert('1')).astype(np.uint8)
annotation_info = pycococreatortools.create_annotation_info(
segmentation_id, image_id, category_info, binary_mask,
image.size, tolerance=2)
if annotation_info is not None:
coco_output["annotations"].append(annotation_info)
segmentation_id = segmentation_id + 1
image_id = image_id + 1
# Store json annotation files
with open(SAVE_PATH_DIR, 'w') as output_json_file:
json.dump(coco_output, output_json_file)
print('ok')
The run json file can be found in the file path of the corresponding setting.
2, The coco label of a single picture and Mask are converted to each other:
2.1 mask conversion of single picture
def objectMultyMask(annFile, classes_names, savePath, type="color"):
"""
From dimension file annFile Extract compliance category classes_names And save it
:param annFile: Label file
:param classes_names: Class alias to be extracted
:param savePath: Mask save path
:param type: Saved color selection,
color use matplot Save as color by default,
gray use cv2 Save as grayscale
:return:
"""
# Get COCO_json data
coco = COCO(annFile)
# Get the id of all the required image data - what is the id of the categories I need
classes_ids = coco.getCatIds(catNms=classes_names)
# Take all picture IDs of the union of all categories
# If you want to intersect, you don't need a loop. Directly enter all categories as parameters to get the pictures contained in all categories
imgIds_list = []
# Loop out the pictures corresponding to each category id and obtain the id number of the picture
for idx in classes_ids:
imgidx = coco.getImgIds(catIds=idx) # Put all the picture IDs of this category into a list
imgIds_list += imgidx
print("search id... ", imgidx)
# Remove duplicate images
imgIds_list = list(set(imgIds_list)) # Merge the same picture id corresponding to multiple categories
# Get the information of all images at one time
image_info_list = coco.loadImgs(imgIds_list)
# Generate a mask for each picture
for imageinfo in image_info_list:
mask_pic = np.zeros(shape=(imageinfo['height'], imageinfo['width']))
for singleClassId in classes_ids:
# Each type of marker generates a mask
singleClassMask = np.zeros(shape=(imageinfo['height'], imageinfo['width']))
# Find the label id of the mask
singleClassAnnId = coco.getAnnIds(imgIds=imageinfo['id'], catIds=[singleClassId], iscrowd=None)
# Extract the segmentation data of the mask
singleClassAnnList = coco.loadAnns(singleClassAnnId)
for singleItem in singleClassAnnList:
# Stack each instance
singleItemMask = coco.annToMask(singleItem)
singleClassMask += singleItemMask
# Finally, this kind of mask information is superimposed on the original image
singleClassMask[singleClassMask > 0] = 1
# The overlapping part uses the mask information of the latter
mask_pic[singleClassMask == 1] = 0
mask_pic = mask_pic + singleClassMask * singleClassId
# Save picture
mask_pic = mask_pic.astype(np.uint8)
file_name = savePath + '/' + imageinfo['file_name'][:-4] + '.png'
if type == "color":
plt.imsave(file_name, mask_pic)
else:
cv2.imwrite(file_name, mask_pic)
print("Saved mask picture: ", file_name)
When saving an image, the value of each pixel represents the category to which the pixel belongs. Therefore, if you need visual display, you can select "color" in the "type" attribute, otherwise select "gary". When running the program directly:
multyMaskGrayDir = "mydata/multyMaskGray"
mkr(multyMaskGrayDir)
objectMultyMask(jsondir, ['date', 'fig', 'hazelnut'], multyMaskGrayDir, type="gray")
multyMaskColorDir = "mydata/multyMaskColor"
mkr(multyMaskColorDir)
objectMultyMask(jsondir, ['date', 'fig', 'hazelnut'], multyMaskColorDir, type="color")
The color display of multi mask is shown in the figure:
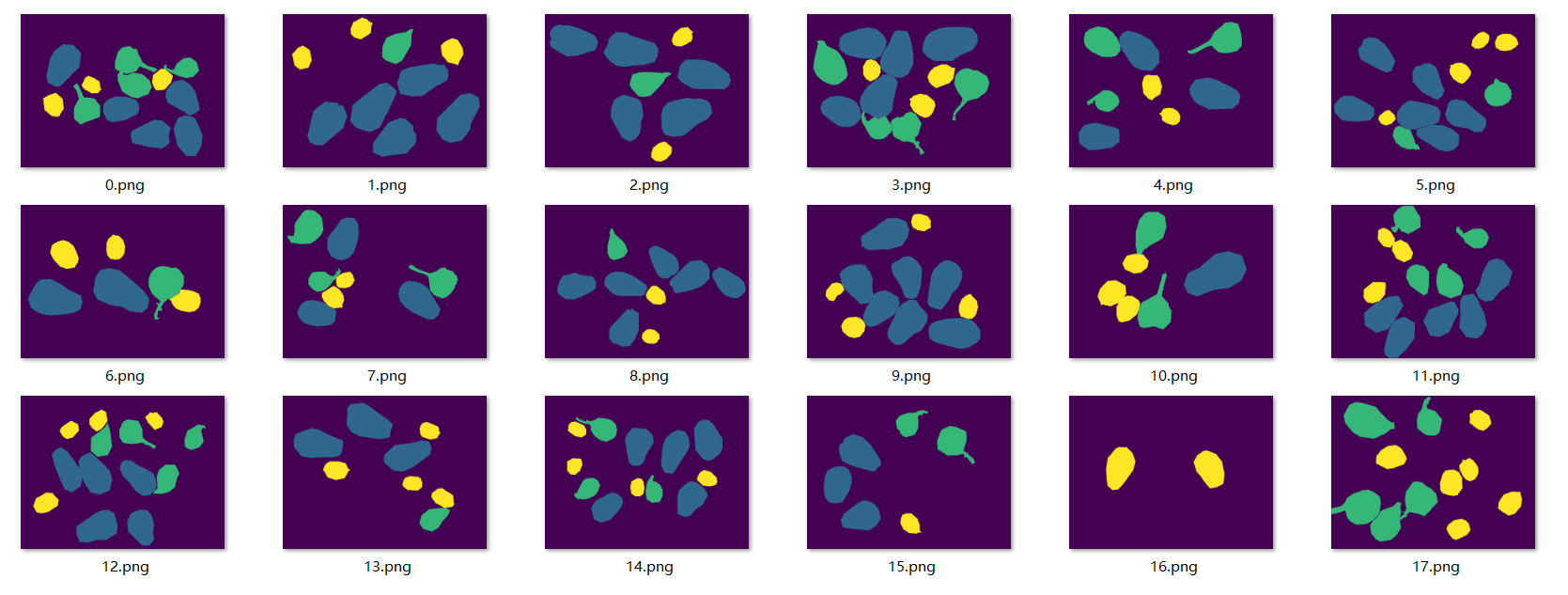
Grayscale image display because the pixel values are 0, 1, 2 and 3, which are displayed in 8-bit images, all are black, which is difficult to recognize by the naked eye, so it will not be displayed here.
2.2 coco annotation file conversion with single picture mask
In the above, we chose the pycococreator tool, which can also be implemented by the tools in cv2
import cv2
import os
import numpy as np
import json
if __name__ == '__main__':
# Initialize the corresponding class alias and id
my_label = {"background": 0,
"date": 1,
"fig": 2,
"hazelnut": 3
}
# Initialize the coco dictionary to be saved
coco = dict()
coco['images'] = []
coco['type'] = 'instances'
coco['annotations'] = []
coco['categories'] = []
# Initialization related variables and Tags
image_set = set()
category_item_id = 0
annotation_id = 0
# 1. Add categories of coco
my_label = sorted(my_label.items(), key=lambda item: item[1])
for val in my_label:
category_item = dict()
category_item['supercategory'] = 'none'
category_item_id = val[1]
if 0 == category_item_id:
continue
category_item['id'] = category_item_id
category_item['name'] = val[0]
coco['categories'].append(category_item)
# 2. Add coco's images
IMAGE_DIR = "data/images/"
ANNOTATION_DIR = "mydata/multyMaskGray/"
SAVE_JSON_DIR = "myjson/multyMaskGray.json"
# Load picture information
imageListFile = os.listdir(IMAGE_DIR)
imageListFile.sort(key=lambda x: int(x[:-4]))
annotationListFile = os.listdir(ANNOTATION_DIR)
annotationListFile.sort(key=lambda x: int(x[:-4]))
assert len(imageListFile) == len(annotationListFile)
for imageId in range(len(imageListFile)):
assert imageListFile[imageId][0:-4] == annotationListFile[imageId][0:-4]
annotationPath = ANNOTATION_DIR + annotationListFile[imageId]
annotationGray = cv2.imread(annotationPath, -1)
if len(annotationGray.shape) == 3:
annotationGray = cv2.cvtColor(annotationGray, cv2.COLOR_BGR2GRAY)
image_item = dict()
image_item['id'] = imageId
image_item['file_name'] = imageListFile[imageId]
image_item['width'] = annotationGray.shape[1] # size['width']
image_item['height'] = annotationGray.shape[0] # size['height']
coco['images'].append(image_item)
image_set.add(imageListFile[imageId])
# 3. Add annotations of coco
for current_category_id in range(1, len(my_label)):
img_bi = np.zeros(annotationGray.shape, dtype='uint8')
img_bi[annotationGray == current_category_id] = 255
my_contours, _ = cv2.findContours(img_bi, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_NONE)
for c in my_contours:
area_t = cv2.contourArea(c)
# The threshold is set here for filtering
if 0 == len(c) or area_t < 20:
continue
L_pt = c
# x,y,w,h
bbox = cv2.boundingRect(L_pt)
x1, y1, w1, h1 = bbox
# If the mark exceeds the limit of the original drawing, ignore it
if x1 < 0 or y1 < 0 or x1 + w1 > annotationGray.shape[1] or y1 + h1 > annotationGray.shape[0]:
continue
seg = []
for val in L_pt:
x = val[0][0]
y = val[0][1]
seg.append(int(x))
seg.append(int(y))
bbox = list(bbox)
annotation_item = dict()
annotation_item['segmentation'] = []
annotation_item['segmentation'].append(seg)
annotation_item['area'] = area_t
annotation_item['iscrowd'] = 0
annotation_item['ignore'] = 0
annotation_item['image_id'] = imageId
annotation_item['bbox'] = bbox
annotation_item['category_id'] = current_category_id
annotation_id += 1
annotation_item['id'] = annotation_id
coco['annotations'].append(annotation_item)
json.dump(coco, open(SAVE_JSON_DIR, 'w'))
print('ok')
The run json file can be found in the file path of the corresponding setting.
3, Display of operation results:
3.1 single instance annotation file training prediction display:
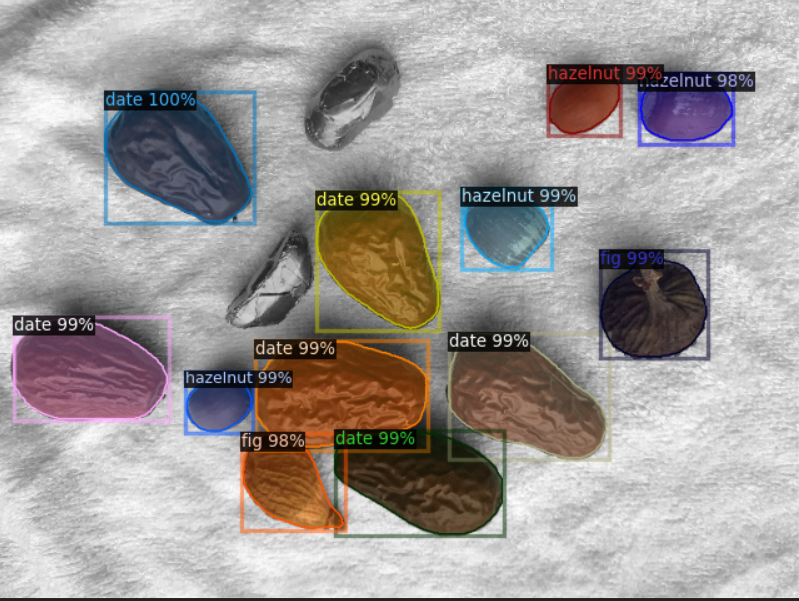
3.2 single picture annotation file training prediction display:
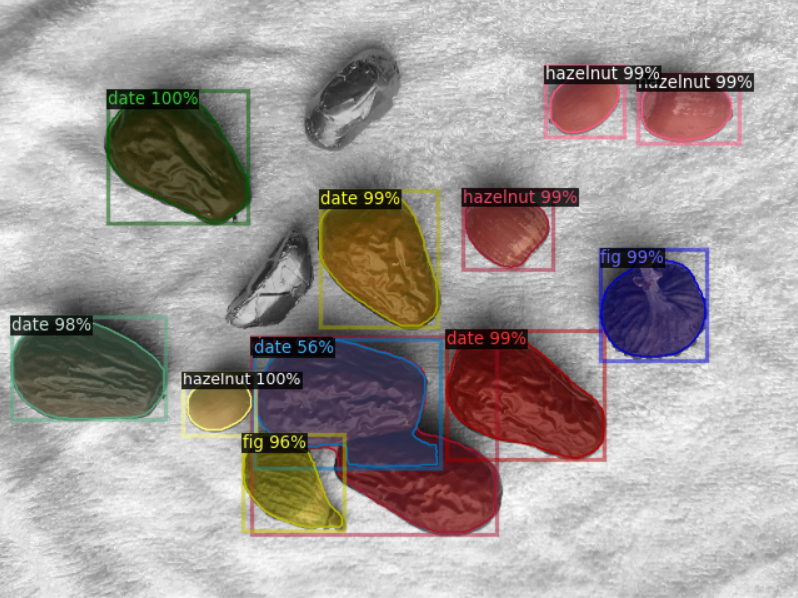
It can be seen that the training effect of single instance annotation file is better than that of single image annotation file. This is because there are two instances connected or blocked in the mask of single image. In this case, it is difficult to separate different instances, so that the training data is trained as a whole. At the same time, after NMS processing, the prediction results will also determine the adjacent objects as a single instance, so the effect is poor.