Classification of ChestRay pneumonia based on KiUnet
This project mainly reproduces the KIUnet network structure and demonstrates how to use the ChestRay pneumonia classification data set.
0. Research motivation
The novel coronavirus pneumonia has become a hot topic in the past. However, novel coronavirus pneumonia is the main criterion of the existing data set.
However, there are many types of pneumonia. This project mainly studies the identification of normal state, viral pneumonia and bacterial pneumonia based on ChestRay data set.
Different from the existing classification network structure of the platform, this project explores the use of split network structure for classification.
Secondly, the KIUnet2D segmentation network is studied and reproduced in this project.
1. Data set introduction
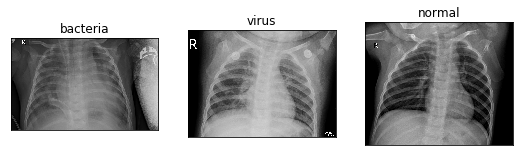
The ChestXRay2017 dataset contains 5856 chest X-ray fluoroscopies. The diagnostic results (i.e. classification labels) are mainly divided into normal and pneumonia, and pneumonia can be subdivided into bacterial pneumonia and viral pneumonia.
Chest X-ray images were selected from pediatric patients aged 1 to 5 years in Guangzhou maternal and child health center. All chest X-ray imaging is part of the patient's routine clinical care.
In order to analyze the chest X-ray images, all chest X-rays were screened to remove all low-quality or unreadable scans, so as to ensure the image quality. Then, two professional doctors grade the image diagnosis. Finally, in order to reduce the image diagnosis error,
The test set was also examined by a third expert.
It is mainly divided into two sub folders: train and test, which are used for model training and testing respectively. In each sub file, it is divided into normal (normal) and pneumonia (pneumonia).
The PNEUMONIA folder contains two types of bacterial and viral PNEUMONIA, which can be distinguished by the naming format of the picture.
Three different data sets are shown as follows: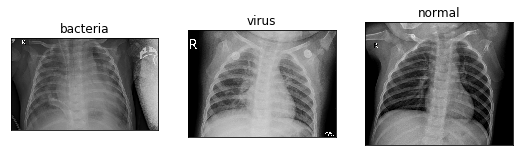
#Decompress the dataset and run it only the first time !unzip -o /home/aistudio/data/data106874/ChestXRay2017.zip -d /home/aistudio/work/
## view picture
import matplotlib.image as mpimg # mpimg is used to read pictures
import matplotlib.pyplot as plt
from PIL import Image
import numpy as np
# Select any three required pictures
_bacteria = 'work/ChestXRay2017/chest_xray/train/PNEUMONIA/person276_bacteria_1296.jpeg'
_virus = 'work/ChestXRay2017/chest_xray/train/PNEUMONIA/person478_virus_975.jpeg'
_normal = 'work/ChestXRay2017/chest_xray/train/NORMAL/NORMAL2-IM-1442-0001.jpeg'
# read
_bacteria = Image.open(_bacteria).convert('RGB')
_virus = Image.open(_virus).convert('RGB')
_normal = Image.open(_normal).convert('RGB')
# mapping
plt.figure(figsize=(9, 5))
plt.subplot(1,3,1),plt.xticks([]),plt.yticks([]),plt.title('bacteria'),plt.imshow(_bacteria)
plt.subplot(1,3,2),plt.xticks([]),plt.yticks([]),plt.title('virus'),plt.imshow(_virus)
plt.subplot(1,3,3),plt.xticks([]),plt.yticks([]),plt.title('normal'),plt.imshow(_normal)
plt.show()
# Install required library functions !pip install paddleseg !pip install opencv-python
import os
import glob
import paddle
from paddle.io import Dataset
import paddleseg.transforms as T
import numpy as np
import random
from PIL import Image
import numpy as np
# Override data read class
class ChestXRayDataset(Dataset):
def __init__(self,mode = 'train',transform =None):
### Read data
rootPath = 'work/ChestXRay2017/chest_xray'
trainPath = os.path.join(rootPath,'train')
testPath = os.path.join(rootPath,'test')
self.transforms = transform
self.mode = mode
if self.mode == 'train':
## Read non pneumonia data
normalTrainPath = os.path.join(trainPath,'NORMAL')
path_ =normalTrainPath + '/*.jpeg' # Match using wildcards
normalTrainList_ = glob.glob(path_)
## Read pneumonia data
normalTrainPath = os.path.join(trainPath,'PNEUMONIA')
# Bacterial pneumonia
path_ =normalTrainPath + '/*bacteria*.jpeg'
bacteriaTrainList_ = glob.glob(path_)
# Viral pneumonia
path_ =normalTrainPath + '/*virus*.jpeg'
virusTrainList_ = glob.glob(path_)
# Add labels 0 [normal], 1 [bacterial pneumonia], 2 [viral pneumonia]
normalTrainList = [[item,0] for item in normalTrainList_]
bacteriaTrainList = [[item,1] for item in bacteriaTrainList_]
virusTrainList = [[item,2] for item in virusTrainList_]
self.jpeg_list = normalTrainList + bacteriaTrainList + virusTrainList
random.shuffle( self.jpeg_list )
else: # test
## Read non pneumonia data
normalTestPath = os.path.join(testPath,'NORMAL')
path_ =normalTestPath + '/*.jpeg'
normalTestList_ = glob.glob(path_)
## Read pneumonia data
normalTestPath = os.path.join(testPath,'PNEUMONIA')
# Bacterial pneumonia
path_ =normalTestPath + '/*bacteria*.jpeg'
bacteriaTestList_ = glob.glob(path_)
# Viral pneumonia
path_ =normalTestPath + '/*virus*.jpeg'
virusTestList_ = glob.glob(path_)
# Add labels 0 [normal], 1 [bacterial pneumonia], 2 [viral pneumonia]
normalTestList = [[item,0] for item in normalTestList_]
bacteriaTestList = [[item,1] for item in bacteriaTestList_]
virusTestList = [[item,2] for item in virusTestList_]
self.jpeg_list = normalTestList + bacteriaTestList + virusTestList
random.shuffle( self.jpeg_list )
def __getitem__(self, index):
pic,label = self.jpeg_list[index]
# Reading ipeg data
data = Image.open(pic).convert('RGB')
#data = data.transpose((2,0,1))
if self.transforms:
data = self.transforms(data)
return data,label
def __len__(self):
return len(self.jpeg_list)
# ## Statistical mean variance is used for data normalization
# import os
# import cv2
# import numpy as np
# import glob
# from tqdm import tqdm
# def getMeanStd(allJpegList_):
# '''
# input: List the paths of all pictures to be calculated
# return: List [means, stdevs]
# '''
# means, stdevs = [], []
# img_dict = {}
# # Using a dictionary is faster than using list and append
# # The reason is that append needs to re apply for an address every time. When the amount of data is large, the subsequent append will be slower and slower. Therefore, it can be changed to dict, and then extract value and turn it into list
# # In this example, the time can be reduced by half, and the more data, the more obvious the acceleration
# for idx,imgs_path in enumerate(allJpegList_):
# img = cv2.cvtColor(cv2.imread(imgs_path), cv2.COLOR_BGR2RGB)
# temp_ = img.reshape(-1,3)
# mean_ = np.expand_dims(np.mean(temp_,0),1)
# std_ = np.expand_dims(np.std(temp_,0),1)
# img_dict[idx] = np.concatenate((mean_,std_),axis=1)
# #Gets the value of the dictionary
# img_list = list(img_dict.values())
# stas_ = np.mean(img_list, axis=0)/255
# return stas_
# # Read training set and test set data
# # The data is RGB data
# path_ ='work/ChestXRay2017/chest_xray/*/*/*.jpeg'
# allJpegList_ = glob.glob(path_)
# statistic_result = getMeanStd(allJpegList_)
# # Statistical results: the first column is the mean of RGB, and the second column is the variance of RGB
# print(statistic_result)
# '''
# [0.48151479 0.22348982]
# [0.48151479 0.22348982]
# [0.48151479 0.22348982]
# '''
2. Introduction of the paper
2.0 KiU-Net: Towards Accurate Segmentation of Biomedical Images using Over-complete Representat
Most medical image segmentation methods using U-Net or its network variants have been successfully applied in most medical scenes.
This "traditional" encoder decoder method has small structure and large error in boundary segmentation.
Literature [1] believes that Unet will pay less attention to shallow features when it pays too much attention to deep features.
In order to overcome this problem, reference [1] proposed a hyperholonomic convolution architecture to convert the input image into a higher dimension in order to suppress the attention of deep receptive fields.
2.1 article highlights
-
The over complete network structure ki net is explored
-
A new network structure Kiu net is proposed by combining incomplete and over complete depth networks
-
It achieves faster convergence speed and better performance in the field of segmentation
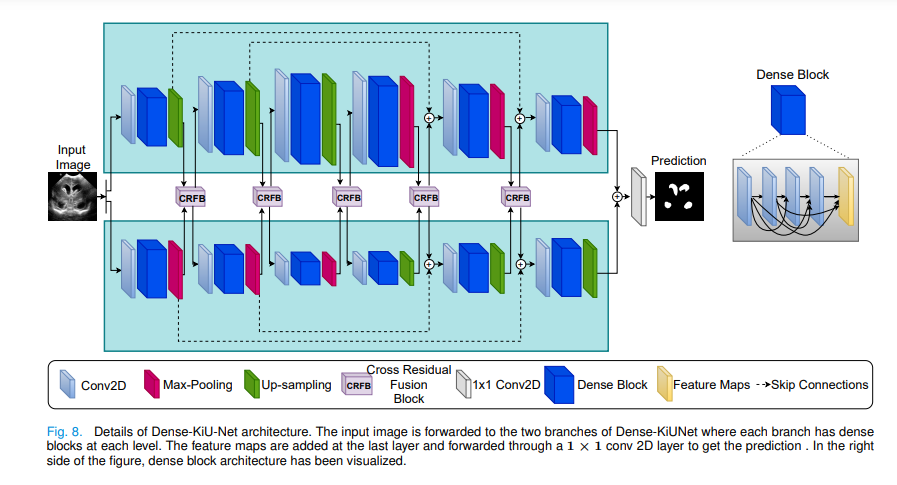
In order to better combine the characteristics of each convolution block, the author proposes a cross residual fusion block.
The characteristic diagram of the same level of the two branches is used as the input at the same time, and then two outputs are obtained as the input of the next level of the two branches.
Add the characteristic diagram of UNet branch to the characteristic diagram of Ki net after passing through the convolution layer and ReLu as the input of the next level of Ki net. alike
The characteristic diagram of Ki net branch is added to the characteristic diagram of UNet after passing through convolution layer and ReLu as the input of the next level of UNet.
Finally, after adding the characteristic images of the two branches, the output segmented image is obtained through a 1x1 convolution. [2]
2.2 network structure
-
The over complete network structure ki net is explored
-
A new network structure Kiu net is proposed by combining incomplete and over complete depth networks
-
It achieves faster convergence speed and better performance in the field of segmentation
2.3 paper results
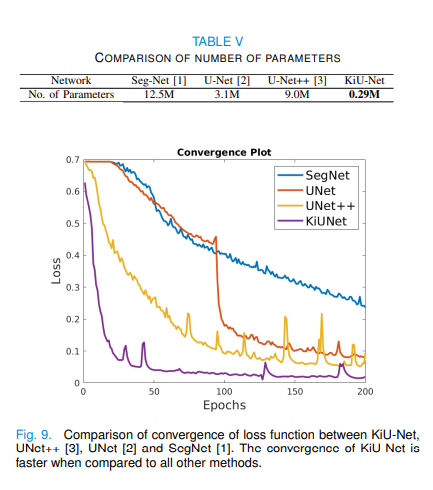
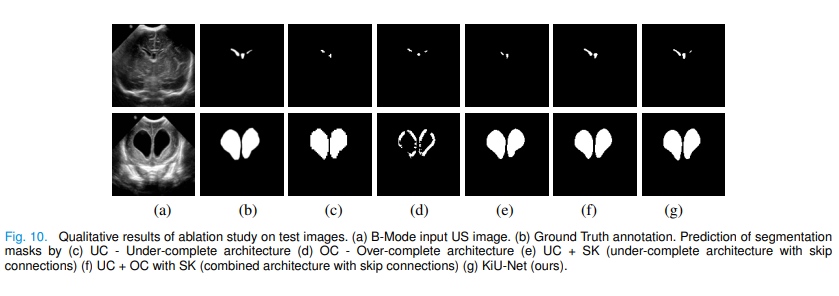
The experimental results show that the training speed is faster, the accuracy is higher, and the model is smaller
The disadvantage is that the actual training takes a long time and requires more memory
2.4 references
[1] Valanarasu J M J, Sindagi V A, Hacihaliloglu I, et al. Kiu-net: Towards accurate segmentation of biomedical images using over-complete representations[C]//International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2020: 363-373.
[2] https://blog.csdn.net/Qy1997/article/details/108356319
# Define KIUnet
import paddle
import paddle.nn as nn
import paddle.nn.functional as F
from paddle.nn import initializer
def init_weights(init_type='kaiming'):
if init_type == 'normal':
return paddle.framework.ParamAttr(initializer=paddle.nn.initializer.Normal())
elif init_type == 'xavier':
return paddle.framework.ParamAttr(initializer=paddle.nn.initializer.XavierNormal())
elif init_type == 'kaiming':
return paddle.framework.ParamAttr(initializer=paddle.nn.initializer.KaimingNormal)
else:
raise NotImplementedError('initialization method [%s] is not implemented' % init_type)
class kiunet(nn.Layer):
def __init__(self ,in_channels = 3, n_classes =3):
super(kiunet,self).__init__()
self.in_channels = in_channels
self.n_class = n_classes
self.encoder1 = nn.Conv2D(self.in_channels, 16, 3, stride=1, padding=1) # First Layer GrayScale Image , change to input channels to 3 in case of RGB
self.en1_bn = nn.BatchNorm(16)
self.encoder2= nn.Conv2D(16, 32, 3, stride=1, padding=1)
self.en2_bn = nn.BatchNorm(32)
self.encoder3= nn.Conv2D(32, 64, 3, stride=1, padding=1)
self.en3_bn = nn.BatchNorm(64)
self.decoder1 = nn.Conv2D(64, 32, 3, stride=1, padding=1)
self.de1_bn = nn.BatchNorm(32)
self.decoder2 = nn.Conv2D(32,16, 3, stride=1, padding=1)
self.de2_bn = nn.BatchNorm(16)
self.decoder3 = nn.Conv2D(16, 8, 3, stride=1, padding=1)
self.de3_bn = nn.BatchNorm(8)
self.decoderf1 = nn.Conv2D(64, 32, 3, stride=1, padding=1)
self.def1_bn = nn.BatchNorm(32)
self.decoderf2= nn.Conv2D(32, 16, 3, stride=1, padding=1)
self.def2_bn = nn.BatchNorm(16)
self.decoderf3 = nn.Conv2D(16, 8, 3, stride=1, padding=1)
self.def3_bn = nn.BatchNorm(8)
self.encoderf1 = nn.Conv2D(in_channels, 16, 3, stride=1, padding=1) # First Layer GrayScale Image , change to input channels to 3 in case of RGB
self.enf1_bn = nn.BatchNorm(16)
self.encoderf2= nn.Conv2D(16, 32, 3, stride=1, padding=1)
self.enf2_bn = nn.BatchNorm(32)
self.encoderf3 = nn.Conv2D(32, 64, 3, stride=1, padding=1)
self.enf3_bn = nn.BatchNorm(64)
self.intere1_1 = nn.Conv2D(16,16,3, stride=1, padding=1)
self.inte1_1bn = nn.BatchNorm(16)
self.intere2_1 = nn.Conv2D(32,32,3, stride=1, padding=1)
self.inte2_1bn = nn.BatchNorm(32)
self.intere3_1 = nn.Conv2D(64,64,3, stride=1, padding=1)
self.inte3_1bn = nn.BatchNorm(64)
self.intere1_2 = nn.Conv2D(16,16,3, stride=1, padding=1)
self.inte1_2bn = nn.BatchNorm(16)
self.intere2_2 = nn.Conv2D(32,32,3, stride=1, padding=1)
self.inte2_2bn = nn.BatchNorm(32)
self.intere3_2 = nn.Conv2D(64,64,3, stride=1, padding=1)
self.inte3_2bn = nn.BatchNorm(64)
self.interd1_1 = nn.Conv2D(32,32,3, stride=1, padding=1)
self.intd1_1bn = nn.BatchNorm(32)
self.interd2_1 = nn.Conv2D(16,16,3, stride=1, padding=1)
self.intd2_1bn = nn.BatchNorm(16)
self.interd3_1 = nn.Conv2D(64,64,3, stride=1, padding=1)
self.intd3_1bn = nn.BatchNorm(64)
self.interd1_2 = nn.Conv2D(32,32,3, stride=1, padding=1)
self.intd1_2bn = nn.BatchNorm(32)
self.interd2_2 = nn.Conv2D(16,16,3, stride=1, padding=1)
self.intd2_2bn = nn.BatchNorm(16)
self.interd3_2 = nn.Conv2D(64,64,3, stride=1, padding=1)
self.intd3_2bn = nn.BatchNorm(64)
self.final = nn.Sequential(
nn.Conv2D(8,self.n_class,1,stride=1,padding=0),
nn.AdaptiveAvgPool2D(output_size=1))
# initialise weights
for m in self.sublayers ():
if isinstance(m, nn.Conv2D):
m.weight_attr = init_weights(init_type='kaiming')
m.bias_attr = init_weights(init_type='kaiming')
elif isinstance(m, nn.BatchNorm):
m.param_attr =init_weights(init_type='kaiming')
m.bias_attr = init_weights(init_type='kaiming')
def forward(self, x):
# input: c * h * w -> 16 * h/2 * w/2
out = F.relu(self.en1_bn(F.max_pool2d(self.encoder1(x),2,2))) #U-Net branch
# c * h * w -> 16 * 2h * 2w
out1 = F.relu(self.enf1_bn(F.interpolate(self.encoderf1(x),scale_factor=(2,2),mode ='bicubic'))) #Ki-Net branch
# 16 * h/2 * w/2
tmp = out
# 16 * 2h * 2w -> 16 * h/2 * w/2
out = paddle.add(out,F.interpolate(F.relu(self.inte1_1bn(self.intere1_1(out1))),scale_factor=(0.25,0.25),mode ='bicubic')) #CRFB
# 16 * h/2 * w/2 -> 16 * 2h * 2w
out1 = paddle.add(out1,F.interpolate(F.relu(self.inte1_2bn(self.intere1_2(tmp))),scale_factor=(4,4),mode ='bicubic')) #CRFB
# 16 * h/2 * w/2
u1 = out #skip conn
# 16 * 2h * 2w
o1 = out1 #skip conn
# 16 * h/2 * w/2 -> 32 * h/4 * w/4
out = F.relu(self.en2_bn(F.max_pool2d(self.encoder2(out),2,2)))
# 16 * 2h * 2w -> 32 * 4h * 4w
out1 = F.relu(self.enf2_bn(F.interpolate(self.encoderf2(out1),scale_factor=(2,2),mode ='bicubic')))
# 32 * h/4 * w/4
tmp = out
# 32 * 4h * 4w -> 32 * h/4 *w/4
out = paddle.add(out,F.interpolate(F.relu(self.inte2_1bn(self.intere2_1(out1))),scale_factor=(0.0625,0.0625),mode ='bicubic'))
# 32 * h/4 * w/4 -> 32 *4h *4w
out1 = paddle.add(out1,F.interpolate(F.relu(self.inte2_2bn(self.intere2_2(tmp))),scale_factor=(16,16),mode ='bicubic'))
# 32 * h/4 *w/4
u2 = out
# 32 *4h *4w
o2 = out1
# 32 * h/4 *w/4 -> 64 * h/8 *w/8
out = F.relu(self.en3_bn(F.max_pool2d(self.encoder3(out),2,2)))
# 32 *4h *4w -> 64 * 8h *8w
out1 = F.relu(self.enf3_bn(F.interpolate(self.encoderf3(out1),scale_factor=(2,2),mode ='bicubic')))
# 64 * h/8 *w/8
tmp = out
# 64 * 8h *8w -> 64 * h/8 * w/8
out = paddle.add(out,F.interpolate(F.relu(self.inte3_1bn(self.intere3_1(out1))),scale_factor=(0.015625,0.015625),mode ='bicubic'))
# 64 * h/8 *w/8 -> 64 * 8h * 8w
out1 = paddle.add(out1,F.interpolate(F.relu(self.inte3_2bn(self.intere3_2(tmp))),scale_factor=(64,64),mode ='bicubic'))
### End of encoder block
### Start Decoder
# 64 * h/8 * w/8 -> 32 * h/4 * w/4
out = F.relu(self.de1_bn(F.interpolate(self.decoder1(out),scale_factor=(2,2),mode ='bicubic'))) #U-NET
# 64 * 8h * 8w -> 32 * 4h * 4w
out1 = F.relu(self.def1_bn(F.max_pool2d(self.decoderf1(out1),2,2))) #Ki-NET
# 32 * h/4 * w/4
tmp = out
# 32 * 4h * 4w -> 32 * h/4 * w/4
out = paddle.add(out,F.interpolate(F.relu(self.intd1_1bn(self.interd1_1(out1))),scale_factor=(0.0625,0.0625),mode ='bicubic'))
# 32 * h/4 * w/4 -> 32 * 4h * 4w
out1 = paddle.add(out1,F.interpolate(F.relu(self.intd1_2bn(self.interd1_2(tmp))),scale_factor=(16,16),mode ='bicubic'))
# 32 * h/4 * w/4
out = paddle.add(out,u2) #skip conn
# 32 * 4h * 4w
out1 = paddle.add(out1,o2) #skip conn
# 32 * h/4 * w/4 -> 16 * h/2 * w/2
out = F.relu(self.de2_bn(F.interpolate(self.decoder2(out),scale_factor=(2,2),mode ='bicubic')))
# 32 * 4h * 4w -> 16 * 2h * 2w
out1 = F.relu(self.def2_bn(F.max_pool2d(self.decoderf2(out1),2,2)))
# 16 * h/2 * w/2
tmp = out
# 16 * 2h * 2w -> 16 * h/2 * w/2
out = paddle.add(out,F.interpolate(F.relu(self.intd2_1bn(self.interd2_1(out1))),scale_factor=(0.25,0.25),mode ='bicubic'))
# 16 * h/2 * w/2 -> 16 * 2h * 2w
out1 = paddle.add(out1,F.interpolate(F.relu(self.intd2_2bn(self.interd2_2(tmp))),scale_factor=(4,4),mode ='bicubic'))
# 16 * h/2 * w/2
out = paddle.add(out,u1)
# 16 * 2h * 2w
out1 = paddle.add(out1,o1)
# 16 * h/2 * w/2 -> 8 * h * w
out = F.relu(self.de3_bn(F.interpolate(self.decoder3(out),scale_factor=(2,2),mode ='bicubic')))
# 16 * 2h * 2w -> 8 * h * w
out1 = F.relu(self.def3_bn(F.max_pool2d(self.decoderf3(out1),2,2)))
# 8 * h * w
out = paddle.add(out,out1) # fusion of both branches
# The last layer activates the function with sigmoid
out = F.sigmoid(self.final(out)) #1*1 conv
return out
# Visual KIunet structure KIunet = kiunet(in_channels = 3, n_classes =3) model = paddle.Model(KIunet) model.summary((2,3, 256, 256))
3. Training code
-
The training time is too long. Only two epochs are trained as instructions
-
Modify the last layer of the network structure and add adaptive avgpool2d to use the network for classification problems.
-
Rich data enhancement methods have been added
model = kiunet(in_channels = 3, n_classes =3)
# Open model training mode
model.train()
# Define the optimization algorithm, use random gradient descent Adam, set the learning rate to 0.001, and the learning rate strategy to stepdelay
scheduler = paddle.optimizer.lr.StepDecay(learning_rate=0.1, step_size=20, gamma=0.1, verbose=False)
optimizer = paddle.optimizer.Adam(learning_rate=scheduler, parameters=model.parameters())
EPOCH_NUM = 2 # Sets the number of outer cycles
BATCH_SIZE = 2 # Set batch size
from paddle.vision import transforms as T
Transforms_train = T.Compose([
T.RandomHorizontalFlip(0.5), # Flip horizontally
T.RandomVerticalFlip(0.5), # Flip vertically
T.RandomRotation(15), # Random rotation
T.Resize(( 128, 128 )), # Resize
T.ColorJitter(0.2, 0.2, 0.2, 0.2),# Randomly adjust brightness, contrast, saturation and hue.
T.Transpose(),
T.Normalize(
[122.78627145, 122.78627145, 122.78627145],
[56.9899041, 56.9899041, 56.9899041]), # Standardization
])
Transforms_test = T.Compose([
T.Resize(( 256, 256 )), # Resize
T.Transpose(),
T.Normalize(
[122.78627145, 122.78627145, 122.78627145],
[56.9899041, 56.9899041, 56.9899041]), # Standardization
])
train_dataset = ChestXRayDataset(mode='train',transform = Transforms_train)
test_dataset = ChestXRayDataset(mode='test',transform = Transforms_test)
# Use the pad io. DataLoader defines the DataLoader object, which is used to load the data generated by the Python generator,
data_loader = paddle.io.DataLoader(train_dataset, batch_size=BATCH_SIZE, shuffle=False)
test_data_loader = paddle.io.DataLoader(test_dataset, batch_size=BATCH_SIZE, shuffle=False)
# Using BCEloss
loss_BCEloss = paddle.nn.BCELoss()
W1227 16:42:41.671527 6593 device_context.cc:447] Please NOTE: device: 0, GPU Compute Capability: 7.0, Driver API Version: 10.1, Runtime API Version: 10.1 W1227 16:42:41.675050 6593 device_context.cc:465] device: 0, cuDNN Version: 7.6.
# Define outer loop
for epoch_id in range(EPOCH_NUM):
# Define inner loop
for iter_id, data in enumerate(data_loader()):
x, y = data # x is the data and y is the label
y = np.squeeze(y)
# Convert tag to onehot encoding
one_hot = paddle.nn.functional.one_hot(y, num_classes=3) #There are three categories
# Convert numpy data into propeller dynamic graph tensor form
x = paddle.to_tensor(x,dtype='float32')
y = paddle.to_tensor(one_hot,dtype='float32')
# Forward calculation
predicts = model(x)
predicts = paddle.squeeze(predicts)
# Calculate loss
loss = loss_BCEloss(predicts, y)
# Clear gradient
optimizer.clear_grad()
# Back propagation
loss.backward()
# Minimize loss and update parameters
optimizer.step()
scheduler.step()
print("epoch: {}, iter: {}, loss is: {}".format(epoch_id+1, iter_id+1, loss.numpy()))
# Save the model parameters with the file name kiunet_model.pdparams
paddle.save(model.state_dict(), 'work/kiunet_model.pdparams')
print("The model is saved successfully, and the model parameters are saved in kiunet_model.pdparams in")
4. Test code
- Calculate average error
import paddle
from sklearn.metrics import accuracy_score
# Model validation
# Clean cache
print("Start test")
# Used to load previously trained model parameters
para_state_dict = paddle.load('work/kiunet_model.pdparams')
model = kiunet(in_channels = 3, n_classes =3)
model.set_dict(para_state_dict)
Error = []
for iter_id, data in enumerate(test_data_loader()):
x, y = data # x is the data and y is the label
# Convert numpy data into propeller dynamic graph tensor form
x = paddle.to_tensor(x,dtype='float32')
# Forward calculation
predicts = model(x)
predicts = paddle.squeeze(predicts)
predicts = predicts.cpu().numpy()
y = y.cpu().numpy()
predLabel = np.argmax(predicts,1)
Error.append(accuracy_score(predLabel, y))
r_id, data in enumerate(test_data_loader()):
x, y = data # x is the data and y is the label
# Convert numpy data into propeller dynamic graph tensor form
x = paddle.to_tensor(x,dtype='float32')
# Forward calculation
predicts = model(x)
predicts = paddle.squeeze(predicts)
predicts = predicts.cpu().numpy()
y = y.cpu().numpy()
predLabel = np.argmax(predicts,1)
Error.append(accuracy_score(predLabel, y))
print("The average positioning error of the test set is:",np.mean(Error))
Start test The average positioning error of the model test set is: 0.38782051282051283
5. Project summary
-
This project mainly implements KIUnet algorithm and realizes the application of modified Unet network in classification task.
-
KIUnet has a long training time and needs a large memory. It is suggested that students interested in the data can modify it into a classification network. There are many excellent classification items with complete annotations on the platform.
-
This project mainly demonstrates the use of the data set and the reproduction of KIUnet network structure. The code annotation is complete and suitable for students interested in segmentation and classification.