introduce
- MobileViT: a lightweight general vision Transformer for mobile devices. According to the author, this is the first lightweight ViT work that can match the performance of lightweight CNN network, representing SOTA! The performance is better than MobileNetV3, CrossViT and other networks.
- Lightweight convolutional neural network (CNN) is a de facto for mobile vision tasks. Their spatial inductive bias enables them to learn to represent with fewer parameters in different visual tasks. However, these networks are spatially local. In order to learn the global representation, a visual Transformer (ViT) based on self attention has been adopted. Unlike CNN, ViT is a "heavyweight". In this paper, we ask the following questions: is it possible to combine the advantages of CNNs and ViTs to build a lightweight and low latency network for mobile vision tasks? To this end, we launched MobileViT, a lightweight universal visual Transformer for mobile devices.
- The structure is also very simple, but it can also achieve a good accuracy performance
- Original paper Download: https://arxiv.org/pdf/2110.02178.pdf
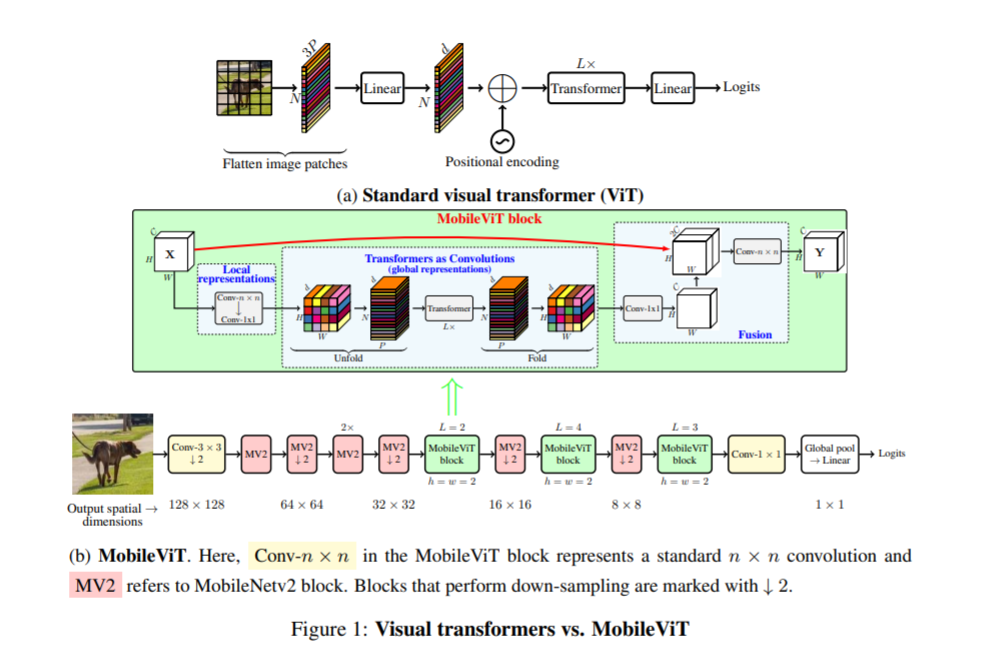
Model architecture
MobileViT is the same as Mobilenet series models. The structure of the model is very simple
- MobileViT brings some new results:
- 1. Better performance: compared with the existing lightweight CNN, the mobilevit model achieves better performance in different mobile vision tasks under the same parameters
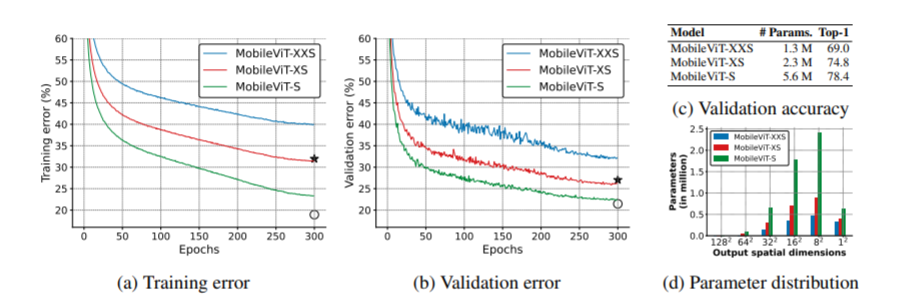
- 2. Better generalization ability: generalization ability refers to the gap between training and evaluation indicators. For two models with similar training indicators, the model with better evaluation indicators is more universal because it can better predict unseen data sets. Compared with CNN, even with extensive data enhancement, its generalization ability is also very poor, and mobilevit shows better generalization ability (as shown in the figure below)
- 3. Better robustness: a good model should be robust to hyperparameters, because tuning these hyperparameters will consume time and resources. Unlike most ViT based models, mobilevit model uses enhancement based training, which is less sensitive to L2 regularization
#!unzip -oq data/data110994/work.zip -d work/
import paddle
paddle.seed(8888)
import numpy as np
from typing import Callable
#Parameter configuration
config_parameters = {
"class_dim": 10, #Classification number
"target_path":"/home/aistudio/work/",
'train_image_dir': '/home/aistudio/work/trainImages',
'eval_image_dir': '/home/aistudio/work/evalImages',
'epochs':20,
'batch_size': 64,
'lr': 0.01
}
#Definition of dataset
class TowerDataset(paddle.io.Dataset):
"""
Step 1: inherit paddle.io.Dataset class
"""
def __init__(self, transforms: Callable, mode: str ='train'):
"""
Step 2: implement the constructor and define the data reading method
"""
super(TowerDataset, self).__init__()
self.mode = mode
self.transforms = transforms
train_image_dir = config_parameters['train_image_dir']
eval_image_dir = config_parameters['eval_image_dir']
train_data_folder = paddle.vision.DatasetFolder(train_image_dir)
eval_data_folder = paddle.vision.DatasetFolder(eval_image_dir)
if self.mode == 'train':
self.data = train_data_folder
elif self.mode == 'eval':
self.data = eval_data_folder
def __getitem__(self, index):
"""
Step 3: Implement__getitem__Methods, defining and specifying index How to obtain data and return a single piece of data (training data, corresponding label)
"""
data = np.array(self.data[index][0]).astype('float32')
data = self.transforms(data)
label = np.array([self.data[index][1]]).astype('int64')
return data, label
def __len__(self):
"""
Step 4: Implement__len__Method to return the total number of data sets
"""
return len(self.data)
from paddle.vision import transforms as T
#Data enhancement
transform_train =T.Compose([T.Resize((256,256)),
#T.RandomVerticalFlip(10),
#T.RandomHorizontalFlip(10),
T.RandomRotation(10),
T.Transpose(),
T.Normalize(mean=[0, 0, 0], # Pixel value normalization
std =[255, 255, 255]), # transforms.ToTensor(), # The transfer operation + (img / 255) and the data structure becomes paddetensor
T.Normalize(mean=[0.50950350, 0.54632660, 0.57409690],# Minus mean divided by standard deviation
std= [0.26059777, 0.26041326, 0.29220656])# Calculation process: output[channel] = (input[channel] - mean[channel]) / std[channel]
])
transform_eval =T.Compose([ T.Resize((256,256)),
T.Transpose(),
T.Normalize(mean=[0, 0, 0], # Pixel value normalization
std =[255, 255, 255]), # transforms.ToTensor(), # The transfer operation + (img / 255) and the data structure becomes paddetensor
T.Normalize(mean=[0.50950350, 0.54632660, 0.57409690],# Minus mean divided by standard deviation
std= [0.26059777, 0.26041326, 0.29220656])# Calculation process: output[channel] = (input[channel] - mean[channel]) / std[channel]
])
train_dataset = TowerDataset(mode='train',transforms=transform_train)
eval_dataset = TowerDataset(mode='eval', transforms=transform_eval )
#Data asynchronous loading
train_loader = paddle.io.DataLoader(train_dataset,
places=paddle.CUDAPlace(0),
batch_size=16,
shuffle=True,
#num_workers=2,
#use_shared_memory=True
)
eval_loader = paddle.io.DataLoader (eval_dataset,
places=paddle.CUDAPlace(0),
batch_size=16,
#num_workers=2,
#use_shared_memory=True
)
print('Training set sample size: {},Sample size of validation set: {}'.format(len(train_loader), len(eval_loader)))
Training set sample size: 1309,Sample size of validation set: 328
code implementation
- The code implementation of the model has actually appeared in the above structure diagram, but it may be difficult to understand because it is too concise
- Another conventional implementation method in the official code is given below. The structure is relatively clear, and some comments are added manually, which is relatively easy to understand
import paddle
import paddle.nn as nn
def conv_1x1_bn(inp, oup):
return nn.Sequential(
nn.Conv2D(inp, oup, 1, 1, 0, bias_attr=False),
nn.BatchNorm2D(oup),
nn.Silu()
)
def conv_nxn_bn(inp, oup, kernal_size=3, stride=1):
return nn.Sequential(
nn.Conv2D(inp, oup, kernal_size, stride, 1, bias_attr=False),
nn.BatchNorm2D(oup),
nn.Silu()
)
class PreNorm(nn.Layer):
def __init__(self, axis, fn):
super().__init__()
self.norm = nn.LayerNorm(axis)
self.fn = fn
def forward(self, x, **kwargs):
return self.fn(self.norm(x), **kwargs)
class FeedForward(nn.Layer):
def __init__(self, axis, hidden_axis, dropout=0.):
super().__init__()
self.net = nn.Sequential(
nn.Linear(axis, hidden_axis),
nn.Silu(),
nn.Dropout(dropout),
nn.Linear(hidden_axis, axis),
nn.Dropout(dropout)
)
def forward(self, x):
return self.net(x)
class Attention(nn.Layer):
def __init__(self, axis, heads=8, axis_head=64, dropout=0.):
super().__init__()
inner_axis = axis_head * heads
project_out = not (heads == 1 and axis_head == axis)
self.heads = heads
self.scale = axis_head ** -0.5
self.attend = nn.Softmax(axis = -1)
self.to_qkv = nn.Linear(axis, inner_axis * 3, bias_attr = False)
self.to_out = nn.Sequential(
nn.Linear(inner_axis, axis),
nn.Dropout(dropout)
) if project_out else nn.Identity()
def forward(self, x):
q,k,v = self.to_qkv(x).chunk(3, axis=-1)
b,p,n,hd = q.shape
b,p,n,hd = k.shape
b,p,n,hd = v.shape
q = q.reshape((b, p, n, self.heads, -1)).transpose((0, 1, 3, 2, 4))
k = k.reshape((b, p, n, self.heads, -1)).transpose((0, 1, 3, 2, 4))
v = v.reshape((b, p, n, self.heads, -1)).transpose((0, 1, 3, 2, 4))
dots = paddle.matmul(q, k.transpose((0, 1, 2, 4, 3))) * self.scale
attn = self.attend(dots)
out = (attn.matmul(v)).transpose((0, 1, 3, 2, 4)).reshape((b, p, n,-1))
return self.to_out(out)
class Transformer(nn.Layer):
def __init__(self, axis, depth, heads, axis_head, mlp_axis, dropout=0.):
super().__init__()
self.layers = nn.LayerList([])
for _ in range(depth):
self.layers.append(nn.LayerList([
PreNorm(axis, Attention(axis, heads, axis_head, dropout)),
PreNorm(axis, FeedForward(axis, mlp_axis, dropout))
]))
def forward(self, x):
for attn, ff in self.layers:
x = attn(x) + x
x = ff(x) + x
return x
class MV2Block(nn.Layer):
def __init__(self, inp, oup, stride=1, expansion=4):
super().__init__()
self.stride = stride
assert stride in [1, 2]
hidden_axis = int(inp * expansion)
self.use_res_connect = self.stride == 1 and inp == oup
if expansion == 1:
self.conv = nn.Sequential(
# dw
nn.Conv2D(hidden_axis, hidden_axis, 3, stride, 1, groups=hidden_axis, bias_attr=False),
nn.BatchNorm2D(hidden_axis),
nn.Silu(),
# pw-linear
nn.Conv2D(hidden_axis, oup, 1, 1, 0, bias_attr=False),
nn.BatchNorm2D(oup),
)
else:
self.conv = nn.Sequential(
# pw
nn.Conv2D(inp, hidden_axis, 1, 1, 0, bias_attr=False),
nn.BatchNorm2D(hidden_axis),
nn.Silu(),
# dw
nn.Conv2D(hidden_axis, hidden_axis, 3, stride, 1, groups=hidden_axis, bias_attr=False),
nn.BatchNorm2D(hidden_axis),
nn.Silu(),
# pw-linear
nn.Conv2D(hidden_axis, oup, 1, 1, 0, bias_attr=False),
nn.BatchNorm2D(oup),
)
def forward(self, x):
if self.use_res_connect:
return x + self.conv(x)
else:
return self.conv(x)
class MobileViTBlock(nn.Layer):
def __init__(self, axis, depth, channel, kernel_size, patch_size, mlp_axis, dropout=0.):
super().__init__()
self.ph, self.pw = patch_size
self.conv1 = conv_nxn_bn(channel, channel, kernel_size)
self.conv2 = conv_1x1_bn(channel, axis)
self.transformer = Transformer(axis, depth, 1, 32, mlp_axis, dropout)
self.conv3 = conv_1x1_bn(axis, channel)
self.conv4 = conv_nxn_bn(2 * channel, channel, kernel_size)
def forward(self, x):
y = x.clone()
# Local representations
x = self.conv1(x)
x = self.conv2(x)
# Global representations
n, c, h, w = x.shape
x = x.transpose((0,3,1,2)).reshape((n,self.ph * self.pw,-1,c))
x = self.transformer(x)
x = x.reshape((n,h,-1,c)).transpose((0,3,1,2))
# Fusion
x = self.conv3(x)
x = paddle.concat((x, y), 1)
x = self.conv4(x)
return x
class MobileViT(nn.Layer):
def __init__(self, image_size, axiss, channels, num_classes, expansion=4, kernel_size=3, patch_size=(2, 2)):
super().__init__()
ih, iw = image_size
ph, pw = patch_size
assert ih % ph == 0 and iw % pw == 0
L = [2, 4, 3]
self.conv1 = conv_nxn_bn(3, channels[0], stride=2)
self.mv2 = nn.LayerList([])
self.mv2.append(MV2Block(channels[0], channels[1], 1, expansion))
self.mv2.append(MV2Block(channels[1], channels[2], 2, expansion))
self.mv2.append(MV2Block(channels[2], channels[3], 1, expansion))
self.mv2.append(MV2Block(channels[2], channels[3], 1, expansion)) # Repeat
self.mv2.append(MV2Block(channels[3], channels[4], 2, expansion))
self.mv2.append(MV2Block(channels[5], channels[6], 2, expansion))
self.mv2.append(MV2Block(channels[7], channels[8], 2, expansion))
self.mvit = nn.LayerList([])
self.mvit.append(MobileViTBlock(axiss[0], L[0], channels[5], kernel_size, patch_size, int(axiss[0]*2)))
self.mvit.append(MobileViTBlock(axiss[1], L[1], channels[7], kernel_size, patch_size, int(axiss[1]*4)))
self.mvit.append(MobileViTBlock(axiss[2], L[2], channels[9], kernel_size, patch_size, int(axiss[2]*4)))
self.conv2 = conv_1x1_bn(channels[-2], channels[-1])
self.pool = nn.AvgPool2D(ih//32, 1)
self.fc = nn.Linear(channels[-1], num_classes, bias_attr=False)
def forward(self, x):
x = self.conv1(x)
x = self.mv2[0](x)
x = self.mv2[1](x)
x = self.mv2[2](x)
x = self.mv2[3](x) # Repeat
x = self.mv2[4](x)
x = self.mvit[0](x)
x = self.mv2[5](x)
x = self.mvit[1](x)
x = self.mv2[6](x)
x = self.mvit[2](x)
x = self.conv2(x)
x = self.pool(x)
x = x.reshape((-1, x.shape[1]))
x = self.fc(x)
return x
def mobilevit_xxs():
axiss = [64, 80, 96]
channels = [16, 16, 24, 24, 48, 48, 64, 64, 80, 80, 320]
return MobileViT((256, 256), axiss, channels, num_classes=1000, expansion=2)
def mobilevit_xs():
axiss = [96, 120, 144]
channels = [16, 32, 48, 48, 64, 64, 80, 80, 96, 96, 384]
return MobileViT((256, 256), axiss, channels, num_classes=1000)
def mobilevit_s():
axiss = [144, 192, 240]
channels = [16, 32, 64, 64, 96, 96, 128, 128, 160, 160, 640]
return MobileViT((256, 256), axiss, channels, num_classes=100)
def count_parameters(model):
return sum(p.numel() for p in model.parameters() if p.requires_grad)
W1114 16:52:06.385679 263 device_context.cc:447] Please NOTE: device: 0, GPU Compute Capability: 7.0, Driver API Version: 10.1, Runtime API Version: 10.1 W1114 16:52:06.390952 263 device_context.cc:465] device: 0, cuDNN Version: 7.6. /opt/conda/envs/python35-paddle120-env/lib/python3.7/site-packages/paddle/nn/layer/norm.py:653: UserWarning: When training, we now always track global mean and variance. "When training, we now always track global mean and variance.") [5, 1000] [5, 1000] [5, 100]
Model test
if __name__ == '__main__':
img = paddle.rand([5, 3, 256, 256])
vit = mobilevit_xxs()
out = vit(img)
print(out.shape)
vit = mobilevit_xs()
out = vit(img)
print(out.shape)
vit = mobilevit_s()
out = vit(img)
print(out.shape)
Instantiation model
model = mobilevit_s() model = paddle.Model(model)
#Optimizer selection
class SaveBestModel(paddle.callbacks.Callback):
def __init__(self, target=0.5, path='work/best_model2', verbose=0):
self.target = target
self.epoch = None
self.path = path
def on_epoch_end(self, epoch, logs=None):
self.epoch = epoch
def on_eval_end(self, logs=None):
if logs.get('acc') > self.target:
self.target = logs.get('acc')
self.model.save(self.path)
print('best acc is {} at epoch {}'.format(self.target, self.epoch))
callback_visualdl = paddle.callbacks.VisualDL(log_dir='work/no_SA')
callback_savebestmodel = SaveBestModel(target=0.5, path='work/best_model1')
callbacks = [callback_visualdl, callback_savebestmodel]
base_lr = config_parameters['lr']
epochs = config_parameters['epochs']
def make_optimizer(parameters=None):
momentum = 0.9
learning_rate= paddle.optimizer.lr.CosineAnnealingDecay(learning_rate=base_lr, T_max=epochs, verbose=False)
weight_decay=paddle.regularizer.L2Decay(0.0001)
optimizer = paddle.optimizer.Momentum(
learning_rate=learning_rate,
momentum=momentum,
weight_decay=weight_decay,
parameters=parameters)
return optimizer
optimizer = make_optimizer(model.parameters())
model.prepare(optimizer,
paddle.nn.CrossEntropyLoss(),
paddle.metric.Accuracy())
model training
model.fit(train_loader,
eval_loader,
epochs=20,
batch_size=1, # Disrupt the sample set
callbacks=callbacks,
verbose=1) # Log display format
Comparative experiment
model_2 = paddle.vision.models.MobileNetV2(num_classes=10 model_2 = paddle.Model(model_2)
#Optimizer selection
class SaveBestModel(paddle.callbacks.Callback):
def __init__(self, target=0.5, path='work/best_model2', verbose=0):
self.target = target
self.epoch = None
self.path = path
def on_epoch_end(self, epoch, logs=None):
self.epoch = epoch
def on_eval_end(self, logs=None):
if logs.get('acc') > self.target:
self.target = logs.get('acc')
self.model.save(self.path)
print('best acc is {} at epoch {}'.format(self.target, self.epoch))
callback_visualdl = paddle.callbacks.VisualDL(log_dir='work/mobilenet_v2')
callback_savebestmodel = SaveBestModel(target=0.5, path='work/best_model')
callbacks = [callback_visualdl, callback_savebestmodel]
base_lr = config_parameters['lr']
epochs = config_parameters['epochs']
def make_optimizer(parameters=None):
momentum = 0.9
learning_rate= paddle.optimizer.lr.CosineAnnealingDecay(learning_rate=base_lr, T_max=epochs, verbose=False)
weight_decay=paddle.regularizer.L2Decay(0.0001)
optimizer = paddle.optimizer.Momentum(
learning_rate=learning_rate,
momentum=momentum,
weight_decay=weight_decay,
parameters=parameters)
return optimizer
optimizer = make_optimizer(model.parameters())
model_2.prepare(optimizer,
paddle.nn.CrossEntropyLoss(),
paddle.metric.Accuracy())
model_2.fit(train_loader,
eval_loader,
epochs=10,
batch_size=1, # Disrupt the sample set
callbacks=callbacks,
```python
model_2.fit(train_loader,
eval_loader,
epochs=10,
batch_size=1, # Disrupt the sample set
callbacks=callbacks,
verbose=1) # Log display format
summary
- The MobileviT model is introduced and implemented, the model alignment and training are realized
- This is a very simple model, but it achieves a good accuracy performance through such a simple model structure. Personally, I think this work is very interesting
Please click here View the basic usage of this environment
Please click here for more detailed instructions.