Medical imaging
Medical image refers to the technology and processing process of obtaining internal tissue image of human body or part of human body in a non-invasive way for medical treatment or medical research.
At present, the main processing methods include: X-ray imaging (X-ray), computed tomography (CT), magnetic resonance imaging (MRI), ultrasonic imaging (ultrasound), positron emission tomography (PET), electroencephalogram (EEG), magnetoencephalogram (MEG), eye tracking, transcranial magnetic wave stimulation (TMS) and other modern imaging technologies, The process of examining parts of the human body that cannot be examined by non-surgical means.
The rise of medical imaging makes the diagnosis of patients have more intuitive evidence. In addition, it also has a great impact on the relevant research in the medical field.
dicom file related
1.1 what is dicom image
DICOM (Digital Imaging and Communications in Medicine), namely medical digital imaging and communication, is an international standard for medical images and related information. It is the most widely used medical image format at present. The common CT, MRI and cardiovascular imaging mostly use docim format.
1.2 what is in DICOM images
doicm mainly stores two aspects of information:
(1) Information about the patient's phi (protected health information)
In short, it is the patient's relevant information (which is protected). For example, name, gender, age, follow-up medical records, etc
(2) Intuitive image information
Part of the information is a certain layer (slice, which will be discussed later) of the scanned patient image. Doctors can open it through a special dicom reader to see the patient's condition. The other part is the relevant equipment information. For example, if the dicom image produced is a layer of the X-ray image scanned by the X-ray machine, dicom will store the relevant equipment information about the X-ray machine.
More specifically, this information can be divided into the following four categories:
(1)Patient
For example, the patient's name, gender, weight, ID, etc. mainly reflect the patient's personal information.
(2)Study
That is, the information recorded about the patient receiving a certain examination, such as examination number, examination date, patient's age at the time of examination, examination location, etc.
(3)Series
Store simple information about the inspection, such as layer thickness, layer spacing, image orientation, etc.
(4)Image
Store detailed information about inspection results (such as a layer of X-ray image), such as image shooting date, image resolution, pixel spacing, etc.
be careful
dicom file ≠ patient image
On the one hand, in the actual examination process, such as CT examination, X-ray and other radioactive light are used to do multi-layer cross-sectional scanning around the human body together with the detector. In other words, the result we get is the cross-sectional images layer by layer. Only when these cross-sectional images are superimposed on the z-axis is a complete medical image. Each dicom stores one layer of images (but not just images). In other words, a patient has multiple dicom files.
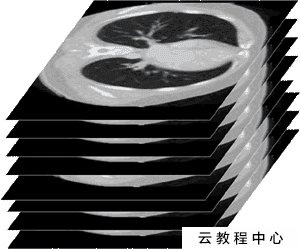
As shown in the figure above, a complete medical image is a three-dimensional image formed by stacking many cross-sectional images on the z-axis. dicom stores only one image and all related information.
On the other hand, dicom files not only store the slices affected by the patient, but also store relevant information (mentioned above).
To sum up, dicom is a file that stores a certain layer of sectional view and related information of medical images taken by patients.
1.3 dicom structure and composition
The internal structure of dicom file is shown in the figure below:
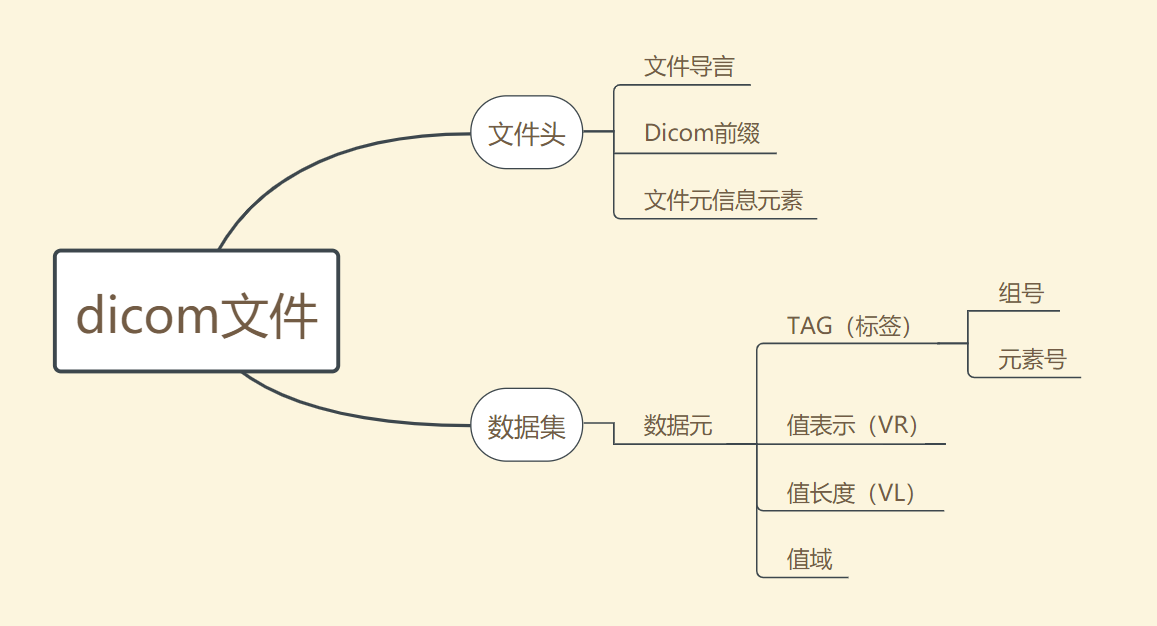
It can be seen that a complete dicom file consists of two parts: file header and data set.
1.3.1 file header
Dicom file header contains relevant information about data sets, mainly including the following parts:
File preamble
Dicom prefix
File information element
1.3.2 data set
Data set is the place where relevant data is stored. The basic unit of data set is data element. The data element consists of four parts:
(1) Tag
That is, the label part is used to identify the specific type of data. It includes two parts: Group number and element number. Here, the code corresponding to Group refers to the category of stored information, that is, indicating that the information is patient,study, series , which of the four categories of image. After determining the Group iu, retrieve the Element to determine the specific information. For example, (00080050) refers to the patient's examination number information.
See Appendix for more specific Tag table.
(2) VR (value representation)
Store the data type of information pointed to by Tag.
(3) VL (data length)
Save the data length of the information pointed to by Tag.
(4) Range of values
Save the specific value of Tag pointing information.
Generally speaking, dicom seems a little troublesome when dealing with it. Therefore, dicom files will be converted to process the converted files. The conversion method used here is to convert dicom files into raw files and mhd files.
raw file related
Raw file, which means "unprocessed file", saves pure pixel information. Often, the images in all dicom files of a patient are extracted and put in a raw file. In other words, a patient corresponds to a raw file, in which the image information of the patient is stored. (it can be understood that different dicom slice images of the patient are stacked together to form a three-dimensional image).
mhd file related
As mentioned above, the dicom file contains some other information besides the slice image. Then, after the file format conversion, the image information is extracted by the raw file, and the non image information is stored in the mhd header file. Simply put, mhd header files store non image information in all dicom files about a patient.
From the above relationship, we can know that raw files and mhd files correspond one-to-one, and their number is less than or equal to (in fact, it must be less than) the number of dicom files.
Corresponding code
2.1 dicom visualization
import matplotlib.patches as patches
import matplotlib.pyplot as plt
import pydicom
from pydicom.data import get_testdata_files
import os
import xlrd
import xlwt
print(__doc__)
path = r'C:\Users\86152.000\Desktop\python\M'
files = os.listdir(path)
for filename in files:
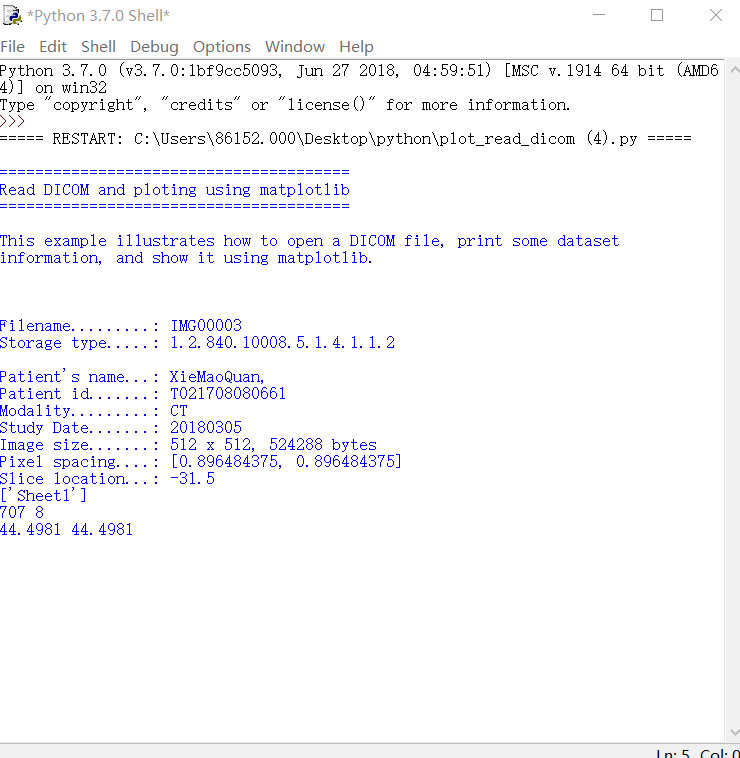
dataset = pydicom.dcmread(filename)
# Normal mode:
print()
print("Filename.........:", filename)
print("Storage type.....:", dataset.SOPClassUID)
print()
pat_name = dataset.PatientName
display_name = pat_name.family_name + ", " + pat_name.given_name
print("Patient's name...:", display_name)
print("Patient id.......:", dataset.PatientID)
print("Modality.........:", dataset.Modality)
print("Study Date.......:", dataset.StudyDate)
if 'PixelData' in dataset:
rows = int(dataset.Rows)
cols = int(dataset.Columns)
print("Image size.......: {rows:d} x {cols:d}, {size:d} bytes".format(
rows=rows, cols=cols, size=len(dataset.PixelData)))
if 'PixelSpacing' in dataset:
print("Pixel spacing....:", dataset.PixelSpacing)
# use .get() if not sure the item exists, and want a default value if missing
print("Slice location...:", dataset.get('SliceLocation', "(missing)"))
def readexcel():
workbook=xlrd.open_workbook(r'C:\Users\86152.000\Desktop\python\annotated_coordinates.xlsx')
print(workbook.sheet_names())
sheet1=workbook.sheet_by_name('Sheet1')
nrows=sheet1.nrows
ncols=sheet1.ncols
print(nrows,ncols)
r=2
c=4
while True:
cellA=sheet1.cell(r,c).value
cellB=sheet1.cell(r,c).value
print(cellA,cellB)
rect=patches.Rectangle((20,30),10,20,linewidth=1,edgecolor='red',facecolor='none')
ax=plt.subplot(1,1,1)
ax.add_patch(rect)
plt.imshow(dataset.pixel_array, cmap=plt.cm.bone)
plt.show()
r=r+1
if r==nrows:
c=c+1
r=2
if c==ncols-2:
print('complete')
break
readexcel()
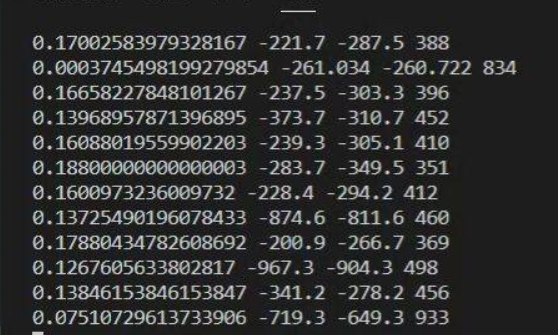
The output is as follows:
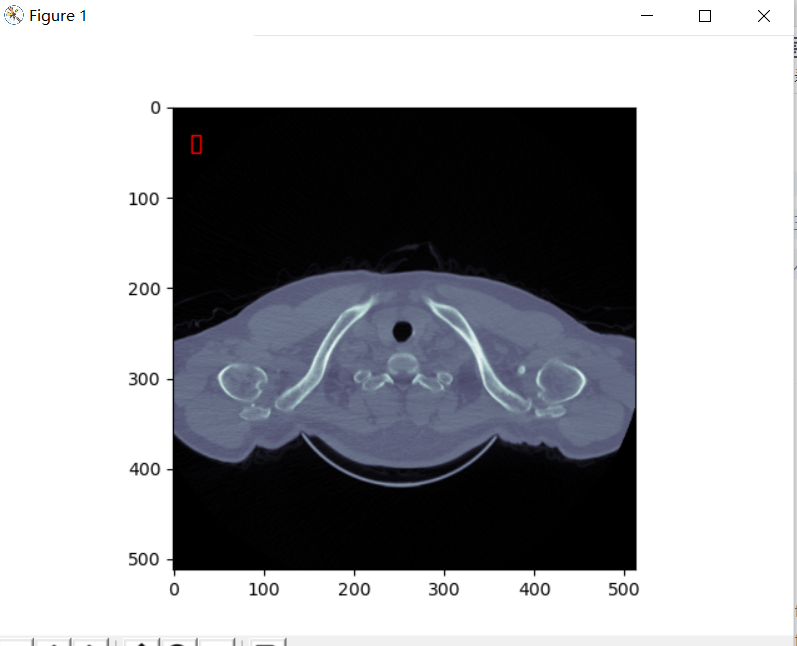
The red box here can be removed and commented in the code. (I wanted to mark dicom data, but the tutor said it should be raw data)
2.2 dicom reads and converts into raw and mhd files
import cv2 import os import pydicom import numpy import SimpleITK
Path and list declarations
A folder in the same directory as the python file stores dicom files. The file path should preferably not contain Chinese
PathDicom = r"/home/Wangling/TuoBaoqiang/python/Test1"
The folder in the same directory as the python file is used to store mhd files and raw files. The file path should preferably not contain Chinese
SaveRawDicom = r"/home/Wangling/TuoBaoqiang/python/Data_Img/test1"
lstFilesDCM= []
lstFilesDCM_0 = []
temp = []
listdicom = []
First level directory under dicom folder - > group one-dimensional list
root_1 = os.listdir(PathDicom)
group = []
for i in range(0,len(root_1)):
group.append(PathDicom + '/' + root_1[i]) #group list
Second level directory under dicom folder - > 000x one-dimensional list
root_2 = []
for j in range(0,len(group)):
root_2. Append (OS. Listdir (Group [J]) # dicom folder under group, root_2 is a two-dimensional list, for example [['0001', '0002', '0003', '0104']]
nameformhd = sum(root_2, []) # will root_ 2 is reduced to a one-dimensional list in case of naming the mhd file at the end of the program
Complete directory of each dicom file - > / group / 000x
for m in range(0,len(group)):
for n in range(0,len(root_2[0]): # suppose that the number of dicom folders in each group is equal, that is, 10. If there is any change, it needs to be modified!
eachdicom = group[m] + '/' + root_2[m][n]
lstFilesDCM_0.append(eachdicom)
print('the dicom filelist - >% s'% lstFilesDCM_0) # print dicom file list
The complete list will be lstfilesdcm_ The dicom file address under 0 is read into listicon
for k in range(0,len(lstFilesDCM_0)):
for dirName, subdirList, fileList in os.walk(lstFilesDCM_0[k]):
for filename in fileList:
if “img” in filename.lower(): # judge whether the file is a dicom file
temp.append(os.path.join(dirName, filename))
listdicom.append(temp) # add to the list
temp = []
print('listicon =% s'% listicon) # print the list of all dicom files loaded
print('the number of listicon is% d '% len (listicon)) # print the number of dicom files to be processed
for filenames in listdicom:
#print('the current processing list is% s'% filenames) # print the dicom list currently being processed
lstFilesDCM = filenames
Step 1: take the last picture as the reference picture and think that all pictures have the same dimension
RefDs = pydicom.read_file(lstFilesDCM[-1],force = True) # Read the last dicom picture RefDs_0 = pydicom.read_file(lstFilesDCM[0],force = True) # Read the first dicom picture, and the third step is standby
Step 2: get the dimension of the 3D picture composed of dicom pictures
ConstPixelDims = (int(RefDs.Rows), int(RefDs.Columns), len(lstFilesDCM)) # ConstPixelDims is a tuple
Step 3: obtain the Spacing in x direction and y direction, and obtain the layer thickness in z direction
RefDs.SliceThickness = abs(RefDs_0.ImagePositionPatient[2]-RefDs.ImagePositionPatient[2])/(len(lstFilesDCM)-1) # Print the value of slicethickness to judge whether the data is correct print(float(RefDs_0.SliceThickness),RefDs.SliceThickness,RefDs_0.ImagePositionPatient[2],RefDs.ImagePositionPatient[2],len(lstFilesDCM)) print(RefDs_0.ImagePositionPatient) print(RefDs.ImagePositionPatient) ConstPixelSpacing = (float(RefDs.PixelSpacing[0]), float(RefDs.PixelSpacing[1]), float(RefDs.SliceThickness))
Step 4: get the origin of the image
Origin = RefDs.ImagePositionPatient
Create a numpy three-dimensional array according to the dimension and set the element type to pixel_array.dtype
ArrayDicom = numpy.zeros(ConstPixelDims, dtype=RefDs.pixel_array.dtype) # array is a numpy array
Step 5: traverse all dicom files, read image data and store it in numpy array
i = 0
for filenameDCM in lstFilesDCM:
ds = pydicom.read_file(filenameDCM)
ArrayDicom[:, :, lstFilesDCM.index(filenameDCM)] = ds.pixel_array
cv2.imwrite("out_" + str(i) + ".png", ArrayDicom[:, :, lstFilesDCM.index(filenameDCM)])
i += 1
Step 6: transpose the numpy array, that is, transform the coordinate axis (x,y,z) into (z,y,x). This is the format of dicom storage file, that is, the first dimension is the Z axis, which is convenient for image stacking
ArrayDicom = numpy.transpose(ArrayDicom, (2, 0, 1))
Step 7: convert the current numpy array into mhd and raw files through SimpleITK
sitk_img = SimpleITK.GetImageFromArray(ArrayDicom, isVector=False) sitk_img.SetSpacing(ConstPixelSpacing) sitk_img.SetOrigin(Origin) SimpleITK.WriteImage(sitk_img,os.path.join(SaveRawDicom,nameformhd[listdicom.index(filenames)]+ ".mhd"))
The output is as follows:
2.3 raw file visualization
import os import SimpleITK as sitk import matplotlib.pyplot as plt
if name=="main":
#Identify Chinese path name
path=r"C:\Users\86152.000\Desktop\python\SaveRaw\0002.mhd"
pwd = os.getcwd()
os.chdir(os.path.dirname(path))
#Read slice image
image = sitk.ReadImage(os.path.basename(path))
image = sitk.GetArrayFromImage(image)
#Display image
for i in range(1,image.shape[0],1):
plt.figure()
plt.imshow(image[i,:,:], cmap='gray')
plt.pause(1)
plt.close()
print(i)
The output is as follows: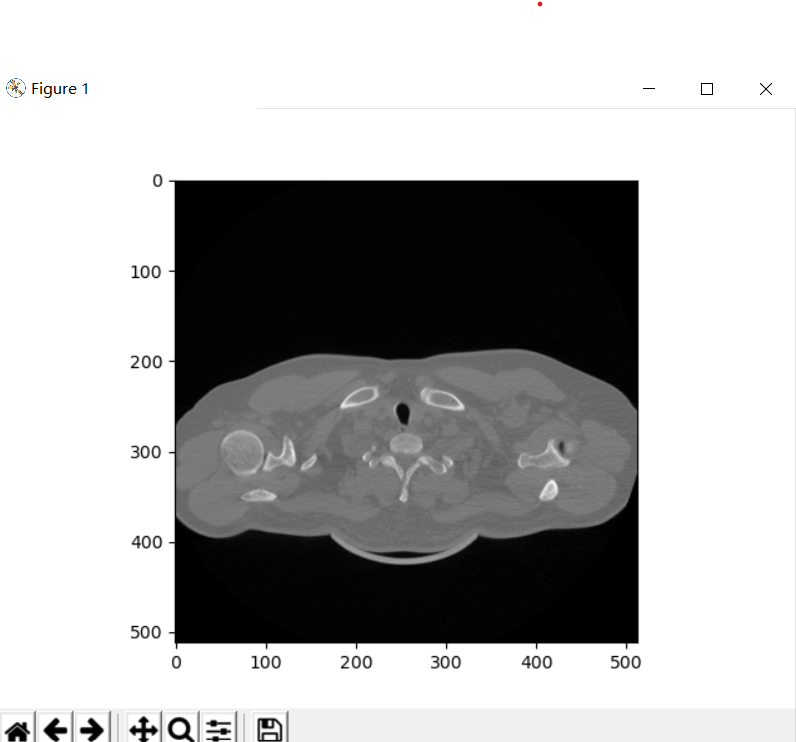
2.4 traversing the specified file
import os import SimpleITK as sitk import matplotlib.pyplot as plt
def show_files(path, all_files):
#First, traverse all files and folders in the current directory
file_list = os.listdir(path)
#Prepare a loop to judge whether each element is a folder or a file. If it is a file, pass the name into the list. If it is a folder, recursion
for file in file_list:
#Using OS path. The join () method obtains the full path name and stores it in cur_path variable, otherwise you can only traverse one layer of directory at a time
cur_path = os.path.join(path, file)
#Determine whether it is a folder
if os.path.isdir(cur_path):
show_files(cur_path, all_files)
else:
#Determine whether the suffix is satisfied
if suffix in cur_path:
all_files.append(cur_path)
return all_files
Operation on the specified file
import os
def show_files(path, all_files):
#First, traverse all files and folders in the current directory
file_list = os.listdir(path)
#Prepare a loop to judge whether each element is a folder or a file. If it is a file, pass the name into the list. If it is a folder, recursion
for file in file_list:
#Using OS path. The join () method obtains the full path name and stores it in cur_path variable, otherwise you can only traverse one layer of directory at a time
cur_path = os.path.join(path, file)
#Determine whether it is a folder
if os.path.isdir(cur_path):
show_files(cur_path, all_files)
else:
#Determine whether the suffix is satisfied
if suffix in cur_path:
all_files.append(cur_path)
return all_files