Hello, I'm classmate K!
Let's move on to the last article 100 deep learning cases | day 51 - target detection algorithm (YOLOv5) (Introduction) After configuring the environment required by YOLOv5, today we try to train our data with YOLOv5. (remember to run through the introductory chapter before starting this tutorial to ensure that other environments are normal)
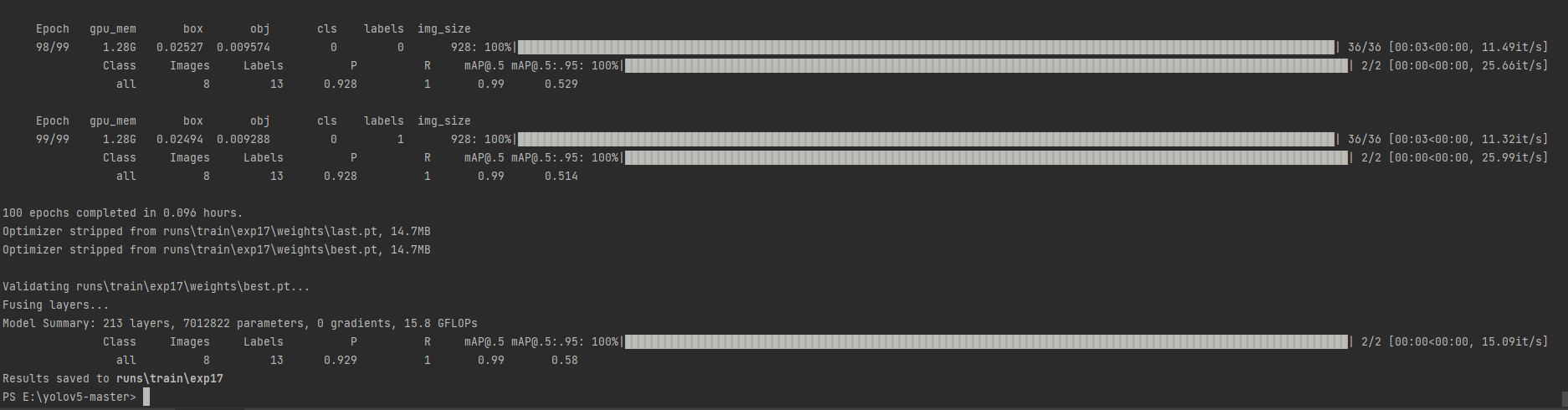
There's a picture and a truth. Let's take a look at my operation results yesterday
[YOLOv5 source address]
1, Prepare your own data
My directory structure is like this
- home directory
- paper_data (create a folder and put the data here)
- Annotations (place our. xml file)
- images (place picture file)
- ImageSets
- Main (four files train.txt, val.txt, test.txt and trainval.txt will be automatically generated in this folder to store the names of training set, verification set and test set pictures)
- paper_data (create a folder and put the data here)
You will see the following directory structure:
The Annotations folder is an xml file. My files are as follows:
My images file is in. png format. The official format is. jpg, but it's not a big problem. Just change the code later (which will be explained later)
2, Run split_train_val.py file
There is a Main subfolder under the ImageSets folder, which stores four files: train.txt, val.txt, test.txt and trainval.txt through split_train_val.py file.
split_ train_ The location of val.py file is as follows:
split_ train_ The content of val.py is as follows:
# coding:utf-8
import os
import random
import argparse
parser = argparse.ArgumentParser()
#The address of the xml file is modified according to its own data. xml is generally stored under Annotations
parser.add_argument('--xml_path', default='Annotations', type=str, help='input xml label path')
#For the division of data sets, select ImageSets/Main under your own data for the address
parser.add_argument('--txt_path', default='ImageSets/Main', type=str, help='output txt label path')
opt = parser.parse_args()
trainval_percent = 1.0
train_percent = 0.9
xmlfilepath = opt.xml_path
txtsavepath = opt.txt_path
total_xml = os.listdir(xmlfilepath)
if not os.path.exists(txtsavepath):
os.makedirs(txtsavepath)
num = len(total_xml)
list_index = range(num)
tv = int(num * trainval_percent)
tr = int(tv * train_percent)
trainval = random.sample(list_index, tv)
train = random.sample(trainval, tr)
file_trainval = open(txtsavepath + '/trainval.txt', 'w')
file_test = open(txtsavepath + '/test.txt', 'w')
file_train = open(txtsavepath + '/train.txt', 'w')
file_val = open(txtsavepath + '/val.txt', 'w')
for i in list_index:
name = total_xml[i][:-4] + '\n'
if i in trainval:
file_trainval.write(name)
if i in train:
file_train.write(name)
else:
file_val.write(name)
else:
file_test.write(name)
file_trainval.close()
file_train.close()
file_val.close()
file_test.close()
Run split_ train_ After the val.py file, you will get four files: train.txt, val.txt, test.txt and trainval.txt. The results are as follows:
3, Generate train.txt, test.txt and val.txt files
Let's first look at the location of the file we want to generate
To get started, what we need now is voc_label.py file, its location is as follows:
voc_ The contents of the label.py file are as follows:
# -*- coding: utf-8 -*-
import xml.etree.ElementTree as ET
import os
from os import getcwd
sets = ['train', 'val', 'test']
classes = ["unripe citrus"] # Change to your own category
abs_path = os.getcwd()
print(abs_path)
def convert(size, box):
dw = 1. / (size[0])
dh = 1. / (size[1])
x = (box[0] + box[1]) / 2.0 - 1
y = (box[2] + box[3]) / 2.0 - 1
w = box[1] - box[0]
h = box[3] - box[2]
x = x * dw
w = w * dw
y = y * dh
h = h * dh
return x, y, w, h
def convert_annotation(image_id):
in_file = open('./Annotations/%s.xml' % (image_id), encoding='UTF-8')
out_file = open('./labels/%s.txt' % (image_id), 'w')
tree = ET.parse(in_file)
root = tree.getroot()
size = root.find('size')
w = int(size.find('width').text)
h = int(size.find('height').text)
for obj in root.iter('object'):
difficult = obj.find('difficult').text
cls = obj.find('name').text
if cls not in classes or int(difficult) == 1:
continue
cls_id = classes.index(cls)
xmlbox = obj.find('bndbox')
b = (float(xmlbox.find('xmin').text), float(xmlbox.find('xmax').text), float(xmlbox.find('ymin').text),
float(xmlbox.find('ymax').text))
b1, b2, b3, b4 = b
# Mark out of range correction
if b2 > w:
b2 = w
if b4 > h:
b4 = h
b = (b1, b2, b3, b4)
bb = convert((w, h), b)
out_file.write(str(cls_id) + " " + " ".join([str(a) for a in bb]) + '\n')
wd = getcwd()
for image_set in sets:
if not os.path.exists('./labels/'):
os.makedirs('./labels/')
image_ids = open('./ImageSets/Main/%s.txt' % (image_set)).read().strip().split()
list_file = open('./%s.txt' % (image_set), 'w')
for image_id in image_ids:
list_file.write(abs_path + '/images/%s.png\n' % (image_id)) # Pay attention to your picture format. If it is. jpg, remember to modify it
convert_annotation(image_id)
list_file.close()
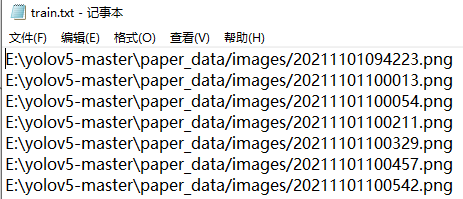
Run voc_label.py file, you will get train.txt, test.txt and val.txt in the screenshot above. The contents of the file are as follows:
4, Create ab.yaml file
I took the file name at will. It can be changed
The location of the ab.yaml file is as follows:
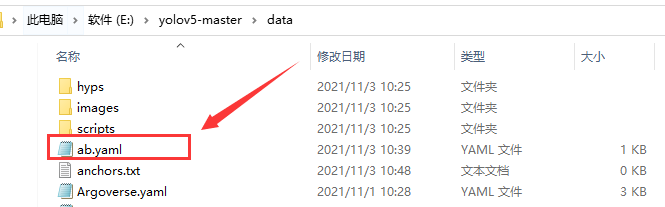
My ab.yaml file reads as follows:
#path: ../datasets/coco # dataset root dir train: ./paper_data/train.txt # train images (relative to 'path') 118287 images val: ./paper_data/val.txt # train images (relative to 'path') 5000 images #test: test-dev2017.txt nc: 1 # number of classes names: ['unripe citrus'] # Change to your own category
5, A priori frame is obtained by clustering
First, we need to prepare the kmeans.py file, which is located in the main directory. Its contents are as follows:
import numpy as np
def iou(box, clusters):
"""
Calculates the Intersection over Union (IoU) between a box and k clusters.
:param box: tuple or array, shifted to the origin (i. e. width and height)
:param clusters: numpy array of shape (k, 2) where k is the number of clusters
:return: numpy array of shape (k, 0) where k is the number of clusters
"""
x = np.minimum(clusters[:, 0], box[0])
y = np.minimum(clusters[:, 1], box[1])
if np.count_nonzero(x == 0) > 0 or np.count_nonzero(y == 0) > 0:
raise ValueError("Box has no area") # If this error is reported, you can change this line to pass
intersection = x * y
box_area = box[0] * box[1]
cluster_area = clusters[:, 0] * clusters[:, 1]
iou_ = intersection / (box_area + cluster_area - intersection)
return iou_
def avg_iou(boxes, clusters):
"""
Calculates the average Intersection over Union (IoU) between a numpy array of boxes and k clusters.
:param boxes: numpy array of shape (r, 2), where r is the number of rows
:param clusters: numpy array of shape (k, 2) where k is the number of clusters
:return: average IoU as a single float
"""
return np.mean([np.max(iou(boxes[i], clusters)) for i in range(boxes.shape[0])])
def translate_boxes(boxes):
"""
Translates all the boxes to the origin.
:param boxes: numpy array of shape (r, 4)
:return: numpy array of shape (r, 2)
"""
new_boxes = boxes.copy()
for row in range(new_boxes.shape[0]):
new_boxes[row][2] = np.abs(new_boxes[row][2] - new_boxes[row][0])
new_boxes[row][3] = np.abs(new_boxes[row][3] - new_boxes[row][1])
return np.delete(new_boxes, [0, 1], axis=1)
def kmeans(boxes, k, dist=np.median):
"""
Calculates k-means clustering with the Intersection over Union (IoU) metric.
:param boxes: numpy array of shape (r, 2), where r is the number of rows
:param k: number of clusters
:param dist: distance function
:return: numpy array of shape (k, 2)
"""
rows = boxes.shape[0]
distances = np.empty((rows, k))
last_clusters = np.zeros((rows,))
np.random.seed()
# the Forgy method will fail if the whole array contains the same rows
clusters = boxes[np.random.choice(rows, k, replace=False)]
while True:
for row in range(rows):
distances[row] = 1 - iou(boxes[row], clusters)
nearest_clusters = np.argmin(distances, axis=1)
if (last_clusters == nearest_clusters).all():
break
for cluster in range(k):
clusters[cluster] = dist(boxes[nearest_clusters == cluster], axis=0)
last_clusters = nearest_clusters
return clusters
if __name__ == '__main__':
a = np.array([[1, 2, 3, 4], [5, 7, 6, 8]])
print(translate_boxes(a))
Then, you need to prepare another one named claim_ Anchors.py, which is also located in the home directory. Its contents are as follows:
# -*- coding: utf-8 -*-
# Find a priori box according to label file
import os
import numpy as np
import xml.etree.cElementTree as et
from kmeans import kmeans, avg_iou
FILE_ROOT = "./paper_data/" # Root path
ANNOTATION_ROOT = "Annotations" # Dataset label folder path
ANNOTATION_PATH = FILE_ROOT + ANNOTATION_ROOT
print(ANNOTATION_PATH)
ANCHORS_TXT_PATH = "./data/anchors.txt"
CLUSTERS = 1 # The number of categories should be modified
CLASS_NAMES = ['unripe citrus'] #Modify to your own category
def load_data(anno_dir, class_names):
xml_names = os.listdir(anno_dir)
boxes = []
for xml_name in xml_names:
xml_pth = os.path.join(anno_dir, xml_name)
tree = et.parse(xml_pth)
width = float(tree.findtext("./size/width"))
height = float(tree.findtext("./size/height"))
for obj in tree.findall("./object"):
cls_name = obj.findtext("name")
if cls_name in class_names:
xmin = float(obj.findtext("bndbox/xmin")) / width
ymin = float(obj.findtext("bndbox/ymin")) / height
xmax = float(obj.findtext("bndbox/xmax")) / width
ymax = float(obj.findtext("bndbox/ymax")) / height
box = [xmax - xmin, ymax - ymin]
boxes.append(box)
else:
continue
return np.array(boxes)
if __name__ == '__main__':
anchors_txt = open(ANCHORS_TXT_PATH, "w")
train_boxes = load_data(ANNOTATION_PATH, CLASS_NAMES)
count = 1
best_accuracy = 0
best_anchors = []
best_ratios = []
for i in range(10): ##### It can be modified, not too large, otherwise it will take a long time
anchors_tmp = []
print(train_boxes)
clusters = kmeans(train_boxes, k=CLUSTERS)
idx = clusters[:, 0].argsort()
clusters = clusters[idx]
# print(clusters)
for j in range(CLUSTERS):
anchor = [round(clusters[j][0] * 640, 2), round(clusters[j][1] * 640, 2)]
anchors_tmp.append(anchor)
print(f"Anchors:{anchor}")
temp_accuracy = avg_iou(train_boxes, clusters) * 100
print("Train_Accuracy:{:.2f}%".format(temp_accuracy))
ratios = np.around(clusters[:, 0] / clusters[:, 1], decimals=2).tolist()
ratios.sort()
print("Ratios:{}".format(ratios))
print(20 * "*" + " {} ".format(count) + 20 * "*")
count += 1
if temp_accuracy > best_accuracy:
best_accuracy = temp_accuracy
best_anchors = anchors_tmp
best_ratios = ratios
anchors_txt.write("Best Accuracy = " + str(round(best_accuracy, 2)) + '%' + "\r\n")
anchors_txt.write("Best Anchors = " + str(best_anchors) + "\r\n")
anchors_txt.write("Best Ratios = " + str(best_ratios))
anchors_txt.close()
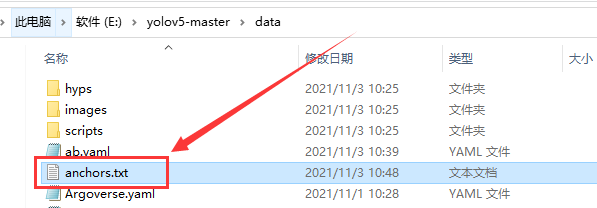
Run claim_ Anchors.py file, we will get anchors.txt file
6, Start training the model with your own dataset
Enter command:
python train.py --img 900 --batch 2 --epoch 100 --data data/ab.yaml --cfg models/yolov5s.yaml --weights weights/yolov5s.pt --device '0'
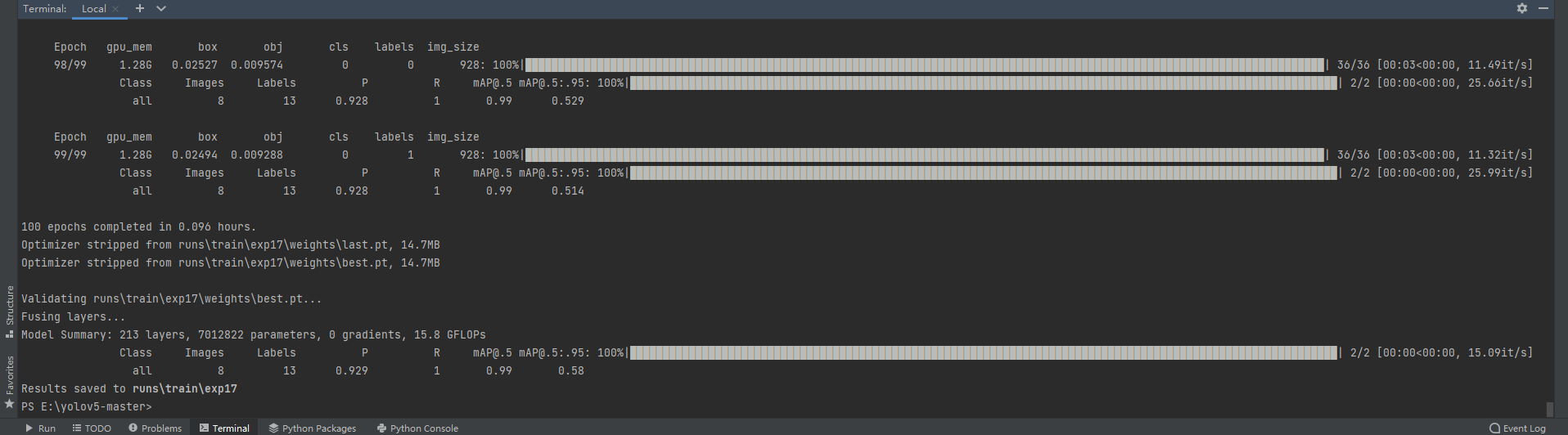
We can directly train our own data set. My final running results are as follows:
If you still have problems that can't be solved, you can add my wechat (mtyjkh#) or leave a message directly below. After seeing it, you will reply to you as soon as possible.